| tags: [ Bivariate Correlations Data Cleaning Imputation Ocular Biometry PCA R ] categories: [Coding Experiments ]
Adding Conjunctival UVAF to dataset
# required packages
require(tidyverse)
require(broom)
require(missMDA)
require(FactoMineR)
require(moments)
require(Hmisc)
require(corrplot)Introduction
Upon inspection of the current data set being used, we realised that there was a variable missing. The variable was a total meausure of conjunctival ultraviolet fluorescence (UVAF). This was included in the original NI Eye Study and therefore, we decided to introduce the variable to this study.
Methods and Results
1. Load phenotype data
phen.data.uvaf <- read.csv('C:/Users/Martha/Documents/Honours/Project/honours.project/Data/NIES_master_database_120815_data_dictionary-2.csv')
head(phen.data.uvaf)## UUID LAB_ID PID SID Ped_ID_.BK_no. NIES NIES_GWAS
## 1 219960 NA 19960 10015417 2438 ES_001 0
## 2 313180 6876 13180 10016420 104 ES_002 0
## 3 314911 6864 14911 10014320 6059 ES_003 0
## 4 111150 1115 NA ES_004 0
## 5 363161 6962 63161 10014308 NA ES_005 0
## 6 110460 1046 NA ES_006 0
## Miles_coreped NIES_DNA DNA_Box DNA_Postition Gender DOB Age.excel
## 1 1 0 NA Male 6/01/1955 53
## 2 1 0 NA Male 21/11/1953 55
## 3 1 0 NA Female 17/04/1955 53
## 4 0 0 NA Male 2/10/1942 66
## 5 0 0 NA Female 4/09/1948 60
## 6 0 0 NA Female 21/10/1938 70
## Height_NIES Weight_NIES Smoker Glaucoma.medications Eye.Colour
## 1 NA NA FALSE Brown
## 2 170 78 FALSE Brown
## 3 180 86 TRUE Hazel / Light Brown
## 4 183 NA TRUE Blue
## 5 NA 69 TRUE Brown
## 6 162 NA TRUE Hazel / Light Brown
## R.Logmar.VA RVA.with.PH L.Logmar.VA LVA.with.PH R.Sph..pre.dilate.
## 1 0.02 0.02 -0.04 -0.04 0.25
## 2 0.10 0.10 0.16 0.10 0.00
## 3 0.00 0.00 0.00 0.00 1.25
## 4 0.30 0.10 0.08 0.08 1.25
## 5 0.00 0.00 -0.10 -0.10 1.25
## 6 0.24 0.16 0.36 0.14 4.00
## R.Cyl..pre.dilate. R.Axis..pre.dilate. L.Sph..pre.dilate.
## 1 0.00 0 0.25
## 2 -0.75 28 -0.50
## 3 -1.25 148 1.25
## 4 -0.25 97 1.50
## 5 -0.25 37 1.25
## 6 -0.50 46 3.75
## L.Cyl..pre.dilate. L.Axis..pre.dilate. IPDprecyclo R.Sph R.Cyl R.Axis
## 1 -0.50 79 NA NA NA NA
## 2 -0.25 164 NA NA NA NA
## 3 -0.25 24 NA NA NA NA
## 4 -0.50 81 NA NA NA NA
## 5 -0.75 164 NA NA NA NA
## 6 -0.75 180 NA NA NA NA
## L.Sph L.Cyl L.Axis IPDpostcyclo Glasses R.Sph.gl R.Cyl.gl R.Axis.gl Add
## 1 NA NA NA NA s gls NA NA NA NA
## 2 NA NA NA NA s gls NA NA NA NA
## 3 NA NA NA NA c gls NA NA NA NA
## 4 NA NA NA NA s gls NA NA NA NA
## 5 NA NA NA NA c gls NA NA NA NA
## 6 NA NA NA NA s gls NA NA NA NA
## L.Sph.gl L.Cyl.gl L.Axis.gl R.K.value.H R.K.Value.H.Axis R.K.value.V
## 1 NA NA NA 42.00 2 43.00
## 2 NA NA NA 41.25 7 42.25
## 3 NA NA NA NA NA NA
## 4 NA NA NA 44.75 5 45.00
## 5 NA NA NA 44.75 0 44.75
## 6 NA NA NA 42.00 8 43.25
## R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## 1 92 42.50 5 43.50
## 2 97 41.50 168 42.00
## 3 NA NA NA NA
## 4 95 45.00 60 45.25
## 5 90 44.25 178 44.75
## 6 98 42.25 177 43.25
## L.K.value.V.Axis Cover.Test.D Lang.score Ocular.Motility IOP.time
## 1 95 NA 550 30/12/1899
## 2 78 NA 550 30/12/1899
## 3 NA NA 550 30/12/1899
## 4 150 NA NA 30/12/1899
## 5 88 NA 550 30/12/1899
## 6 87 NA 550 30/12/1899
## R.Pachimetry L.Pachimetry R.Axial.Length L.Axial.Length R.AC.Depth
## 1 532 554 24.31 24.10 3.09
## 2 608 612 25.02 25.21 3.38
## 3 507 510 22.78 22.80 3.40
## 4 560 559 23.02 22.98 3.00
## 5 556 562 21.75 22.04 2.60
## 6 498 501 23.06 23.17 2.94
## L.AC.Depth R.IOP.mmHg L.IOP.mmHg
## 1 3.03 14 14
## 2 3.92 16 15
## 3 3.45 26 22
## 4 2.85 14 14
## 5 2.53 22 21
## 6 3.04 18 20
## Anterior.segment Pterygium
## 1 OD NAD, OS mild cortical cataract FALSE
## 2 OD NAD, OS flecks on posterior capsule 'present since age 16.' TRUE
## 3 FALSE
## 4 cortical cataract OU (R>L) FALSE
## 5 OD pterygium, OS NAD TRUE
## 6 NAD OU FALSE
## R.Disc.size L.Disc.size R.CDR L.CDR FDT.MD_RE FDT.MD_LE FDT.PSD_RE
## 1 L L 0.9 0.9 6.08 5.07 2.050
## 2 L L 0.9 0.7 1.21 -0.72 1.600
## 3 L L 0.7 0.7 2.33 2.28 2.653
## 4 NR NR 0.2 0.2 NA NA NA
## 5 M M 0.3 0.3 NA NA NA
## 6 L L 0.6 0.6 NA NA NA
## FDT.PSD_LE
## 1 2.09
## 2 2.78
## 3 2.92
## 4 NA
## 5 NA
## 6 NA
## Comments
## 1 Suspicious discs (OU)
## 2 Suspicious discs. Fields OK, but left 1 small defect. Check in 1-2 years.
## 3 OHT but field defect on R not classic. Need a 6-12/12 review no axial length
## 4
## 5 No diabetic retinopathy
## 6
## R.L.handed R.L.eye.dominance Other.disease UV.questionnaire_NIES.code
## 1 R 1
## 2 L 2
## 3 L 3
## 4 R 4
## 5 R 5
## 6 R 6
## UV.questionnaire_First.name Maiden.name UV.questionnaire_Surname
## 1 NA NA NA
## 2 NA NA NA
## 3 NA NA NA
## 4 NA NA NA
## 5 NA NA NA
## 6 NA NA NA
## UV.questionnaire_Age UV.questionnaire_Sex Lived.in.Norfolk.Island
## 1 52 M 37
## 2 53 M 46
## 3 52 F 21
## 4 65 M 28
## 5 59 F 16
## 6 68 F 16
## Location. From.age.in.years Duration.outside.Norfolk.Island
## 1 SA, WA, NSW 18 15
## 2 PNG NA 7
## 3 Australia/ PNG 0 31
## 4 0 37
## 5 0 43
## 6 QLD 0 52
## Hair.colour Burns.Tans Sunburn.and.pain Time.outside
## 1 Black burns then tans more than 10 times greater than 3/4 day
## 2 Brown never burns but tans 2-10 times none
## 3 Brown burns not tan more than 10 times less than 1/4day
## 4 Brown burns not tan 2-10 times cannot judge
## 5 Brown burns then tans once less than 1/4day
## 6 Brown burns not tan 2-10 times less than 1/4day
## Outdoors.and.hats Outdoors.and.sunglasses Winter..Indoors.and.Outdoors
## 1 always 1/2 of the time mostly outdoors
## 2 1/2 of the time seldom 1/2 and 1/2
## 3 1/2 of the time usually 1/2 and 1/2
## 4 always seldom 1/2 and 1/2
## 5 usually always mostly indoors
## 6 usually usually mostly indoors
## At.school Feel.colder.than.people Arc.Welding
## 1 yes, sunglasses only seldom Yes
## 2 neither 1/2 of the time Yes
## 3 neither 1/2 of the time No
## 4 neither seldom Yes
## 5 neither seldom No
## 6 neither seldom No
## If.yes..eye.protection If.yes..flash.burn. Reason.for.sunglasses
## 1 No No protection from eye disease
## 2 No No glare
## 3 glare
## 4 No No glare
## 5 driving
## 6 glare
## Do.not.wear.sunglasses X0531_niesid totaluvafmm NIESMatch GLC_nies_code
## 1 Inconvenient 1 0.0 TRUE NA
## 2 Decrease vision 2 74.4 TRUE NA
## 3 3 14.6 TRUE NA
## 4 Not necessary 4 1.9 TRUE NA
## 5 NA NA TRUE NA
## 6 Prescription glasses 6 1.0 TRUE NA
## Glaucoma Diagnosis.GLC.known.previous. PIT GLC_griffith_code
## 1 NA
## 2 NA
## 3 NA
## 4 NA
## 5 NA
## 6 NA2. Perform summary statistics
summary.uvaf <- summary(phen.data.uvaf)
summary.uvaf## UUID LAB_ID PID SID
## Min. :110150 Min. :1001 :391 :391
## 1st Qu.:310050 1st Qu.:1181 10050 : 1 10012610: 1
## Median :327190 Median :2509 10071 : 1 10014214: 1
## Mean :323827 Mean :3644 10121 : 1 10014215: 1
## 3rd Qu.:400396 3rd Qu.:6858 10280 : 1 10014216: 1
## Max. :400805 Max. :7000 10291 : 1 10014218: 1
## NA's :449 (Other):405 (Other) :405
## Ped_ID_.BK_no. NIES NIES_GWAS Miles_coreped
## Min. : 2 ES_001 : 1 Min. :0.0000 Min. :0.0000
## 1st Qu.: 812 ES_002 : 1 1st Qu.:0.0000 1st Qu.:0.0000
## Median :1921 ES_003 : 1 Median :0.0000 Median :1.0000
## Mean :2239 ES_004 : 1 Mean :0.1498 Mean :0.5468
## 3rd Qu.:3449 ES_005 : 1 3rd Qu.:0.0000 3rd Qu.:1.0000
## Max. :7080 ES_006 : 1 Max. :1.0000 Max. :1.0000
## NA's :288 (Other):795
## NIES_DNA DNA_Box DNA_Postition Gender
## Min. :0.0000 Min. :1.000 :566 Female:443
## 1st Qu.:0.0000 1st Qu.:1.000 A1 : 4 Male :358
## Median :0.0000 Median :2.000 A2 : 4
## Mean :0.2934 Mean :2.315 A4 : 4
## 3rd Qu.:1.0000 3rd Qu.:3.000 A5 : 4
## Max. :1.0000 Max. :4.000 A6 : 4
## NA's :566 (Other):215
## DOB Age.excel Height_NIES Weight_NIES
## 11/12/1995: 2 Min. : 8.00 Min. :127.0 Min. : 31.00
## 12/03/1965: 2 1st Qu.:42.00 1st Qu.:162.4 1st Qu.: 65.00
## 15/11/1990: 2 Median :55.00 Median :170.0 Median : 75.00
## 18/04/1958: 2 Mean :53.27 Mean :169.7 Mean : 76.84
## 19/01/1971: 2 3rd Qu.:66.00 3rd Qu.:177.6 3rd Qu.: 86.00
## 19/12/1995: 2 Max. :91.00 Max. :195.0 Max. :140.00
## (Other) :789 NA's :137 NA's :163
## Smoker Glaucoma.medications Eye.Colour
## Mode :logical :769 Brown :249
## FALSE:359 Xalatan : 6 Blue :242
## TRUE :442 Timoptol: 3 Hazel / Light Brown: 85
## xalatan : 3 Green : 81
## Xalacom : 2 : 64
## Alphagan: 1 Hazel : 33
## (Other) : 17 (Other) : 47
## R.Logmar.VA RVA.with.PH L.Logmar.VA
## Min. :-0.30000 Min. :-0.24000 Min. :-0.30000
## 1st Qu.:-0.10000 1st Qu.:-0.08000 1st Qu.:-0.10000
## Median : 0.02000 Median : 0.02000 Median : 0.02000
## Mean : 0.06266 Mean : 0.04614 Mean : 0.05989
## 3rd Qu.: 0.12000 3rd Qu.: 0.10000 3rd Qu.: 0.12000
## Max. : 2.00000 Max. : 2.00000 Max. : 1.70000
## NA's :3 NA's :210 NA's :6
## LVA.with.PH R.Sph..pre.dilate. R.Cyl..pre.dilate.
## Min. :-0.2600 Min. :-8.0000 Min. :-11.0000
## 1st Qu.:-0.0800 1st Qu.: 0.0000 1st Qu.: -0.7500
## Median : 0.0400 Median : 0.5000 Median : -0.5000
## Mean : 0.0529 Mean : 0.5411 Mean : -0.6389
## 3rd Qu.: 0.1000 3rd Qu.: 1.2500 3rd Qu.: -0.2500
## Max. : 1.0600 Max. : 8.5000 Max. : 0.0000
## NA's :214 NA's :11 NA's :11
## R.Axis..pre.dilate. L.Sph..pre.dilate. L.Cyl..pre.dilate.
## Min. : 0.00 Min. :-9.5000 Min. :-9.7500
## 1st Qu.: 25.00 1st Qu.: 0.0000 1st Qu.:-0.7500
## Median : 85.00 Median : 0.5000 Median :-0.5000
## Mean : 80.22 Mean : 0.5781 Mean :-0.6403
## 3rd Qu.:119.75 3rd Qu.: 1.2500 3rd Qu.:-0.2500
## Max. :180.00 Max. : 8.0000 Max. : 0.0000
## NA's :11 NA's :10 NA's :10
## L.Axis..pre.dilate. IPDprecyclo R.Sph R.Cyl
## Min. : 0.00 Min. : 2.00 Min. :0.2500 Min. :-1.0000
## 1st Qu.: 12.50 1st Qu.:57.50 1st Qu.:0.2500 1st Qu.:-0.6875
## Median : 70.00 Median :59.50 Median :0.3750 Median :-0.3750
## Mean : 71.96 Mean :56.36 Mean :0.5000 Mean :-0.4583
## 3rd Qu.:112.00 3rd Qu.:61.50 3rd Qu.:0.6875 3rd Qu.:-0.2500
## Max. :180.00 Max. :69.00 Max. :1.0000 Max. : 0.0000
## NA's :10 NA's :787 NA's :795 NA's :795
## R.Axis L.Sph L.Cyl L.Axis
## Min. : 0.00 Min. :0.2500 Min. :-1.0000 Min. : 36.00
## 1st Qu.: 50.25 1st Qu.:0.3125 1st Qu.:-0.6875 1st Qu.: 78.75
## Median : 86.00 Median :0.5000 Median :-0.3750 Median :112.00
## Mean : 71.50 Mean :0.5417 Mean :-0.5000 Mean :100.67
## 3rd Qu.: 97.75 3rd Qu.:0.6875 3rd Qu.:-0.2500 3rd Qu.:125.00
## Max. :117.00 Max. :1.0000 Max. :-0.2500 Max. :147.00
## NA's :795 NA's :795 NA's :795 NA's :795
## IPDpostcyclo Glasses R.Sph.gl R.Cyl.gl
## Min. : 0.0000 : 3 Min. :-0.500 Min. :-2.000
## 1st Qu.: 0.0000 c CL's: 2 1st Qu.: 0.250 1st Qu.:-1.375
## Median : 0.0000 c gls :234 Median : 1.000 Median :-0.750
## Mean : 0.9776 cCl's : 1 Mean : 2.250 Mean :-1.167
## 3rd Qu.: 0.0000 cCL's : 1 3rd Qu.: 3.625 3rd Qu.:-0.750
## Max. :64.0000 s gls :560 Max. : 6.250 Max. :-0.750
## NA's :489 NA's :798 NA's :798
## R.Axis.gl Add L.Sph.gl L.Cyl.gl
## Min. : 15.00 Mode:logical Min. :-0.2500 Min. :-1.0000
## 1st Qu.: 67.50 NA's:801 1st Qu.: 0.1250 1st Qu.:-0.8750
## Median :120.00 Median : 0.5000 Median :-0.7500
## Mean : 90.67 Mean : 0.6667 Mean :-0.6667
## 3rd Qu.:128.50 3rd Qu.: 1.1250 3rd Qu.:-0.5000
## Max. :137.00 Max. : 1.7500 Max. :-0.2500
## NA's :798 NA's :798 NA's :798
## L.Axis.gl R.K.value.H R.K.Value.H.Axis R.K.value.V
## Min. : 41.00 Min. :36.00 Min. : 0.0 Min. :37.00
## 1st Qu.: 54.50 1st Qu.:42.00 1st Qu.: 42.0 1st Qu.:42.75
## Median : 68.00 Median :43.00 Median : 92.0 Median :43.75
## Mean : 91.33 Mean :42.94 Mean : 93.5 Mean :43.77
## 3rd Qu.:116.50 3rd Qu.:44.00 3rd Qu.:155.0 3rd Qu.:44.75
## Max. :165.00 Max. :48.25 Max. :180.0 Max. :55.00
## NA's :798 NA's :17 NA's :17 NA's :17
## R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## Min. : 1.00 Min. :32.75 Min. : 0.00 Min. :39.00
## 1st Qu.: 61.00 1st Qu.:42.00 1st Qu.: 26.00 1st Qu.:42.75
## Median : 89.00 Median :43.00 Median : 89.00 Median :43.75
## Mean : 90.69 Mean :42.94 Mean : 88.33 Mean :43.84
## 3rd Qu.:126.25 3rd Qu.:44.00 3rd Qu.:147.00 3rd Qu.:44.75
## Max. :189.00 Max. :47.25 Max. :180.00 Max. :52.00
## NA's :17 NA's :16 NA's :16 NA's :16
## L.K.value.V.Axis Cover.Test.D Lang.score Ocular.Motility
## Min. : 0.00 Mode:logical Min. : -99.0 :726
## 1st Qu.: 65.00 NA's:801 1st Qu.: 550.0 CT N & D Straight: 6
## Median : 90.00 Median : 550.0 lang 0/3 : 6
## Mean : 92.25 Mean : 560.3 Lang 0/3 : 3
## 3rd Qu.:121.00 3rd Qu.: 550.0 LANG 0/3 : 2
## Max. :180.00 Max. :1600.0 ? CN IV palsy : 1
## NA's :16 NA's :98 (Other) : 57
## IOP.time R.Pachimetry L.Pachimetry R.Axial.Length
## : 17 Min. :428.0 Min. :424.0 Min. :20.95
## 30/12/1899:784 1st Qu.:527.0 1st Qu.:526.0 1st Qu.:22.90
## Median :546.0 Median :547.0 Median :23.49
## Mean :546.4 Mean :546.7 Mean :23.56
## 3rd Qu.:570.0 3rd Qu.:569.0 3rd Qu.:24.09
## Max. :656.0 Max. :658.0 Max. :27.66
## NA's :15 NA's :17 NA's :16
## L.Axial.Length R.AC.Depth L.AC.Depth R.IOP.mmHg
## Min. :21.29 Min. :2.090 Min. :2.000 Min. : 6.00
## 1st Qu.:22.87 1st Qu.:3.060 1st Qu.:3.045 1st Qu.:14.00
## Median :23.46 Median :3.320 Median :3.290 Median :16.00
## Mean :23.53 Mean :3.327 Mean :3.314 Mean :15.89
## 3rd Qu.:24.10 3rd Qu.:3.560 3rd Qu.:3.560 3rd Qu.:18.00
## Max. :34.43 Max. :4.950 Max. :5.130 Max. :28.00
## NA's :16 NA's :14 NA's :14 NA's :3
## L.IOP.mmHg Anterior.segment Pterygium R.Disc.size
## Min. : 8.00 NAD OU :271 Mode :logical : 41
## 1st Qu.:14.00 NAD :174 FALSE:684 L : 82
## Median :16.00 IOL OU : 20 TRUE :117 M :493
## Mean :16.06 pterygium OU : 20 NR:112
## 3rd Qu.:18.00 : 18 S : 73
## Max. :33.00 pseudophakia OU: 13
## NA's :4 (Other) :285
## L.Disc.size R.CDR L.CDR FDT.MD_RE
## : 43 Min. :0.000 Min. :0.0000 Min. :-17.790
## L : 82 1st Qu.:0.300 1st Qu.:0.3000 1st Qu.: -2.240
## M :495 Median :0.400 Median :0.4000 Median : 0.000
## NR:109 Mean :0.402 Mean :0.3988 Mean : -1.020
## S : 72 3rd Qu.:0.500 3rd Qu.:0.5000 3rd Qu.: 1.657
## Max. :0.990 Max. :0.9000 Max. : 6.480
## NA's :21 NA's :19 NA's :693
## FDT.MD_LE FDT.PSD_RE FDT.PSD_LE
## Min. :-17.120 Min. :-3.070 Min. : 2.010
## 1st Qu.: -1.855 1st Qu.: 2.540 1st Qu.: 2.638
## Median : -0.220 Median : 3.100 Median : 3.020
## Mean : -1.094 Mean : 3.566 Mean : 3.663
## 3rd Qu.: 1.660 3rd Qu.: 4.000 3rd Qu.: 3.940
## Max. : 5.200 Max. :12.030 Max. :14.720
## NA's :694 NA's :696 NA's :697
## Comments R.L.handed R.L.eye.dominance
## :534 : 26 :787
## LANG 0/3 : 13 \\: 1 LE: 2
## Fields OK. : 2 L : 56 RE: 12
## LANG 0/3. : 2 NR: 95
## No diabetic retinopathy: 2 R :623
## No K's : 2
## (Other) :246
## Other.disease
## :731
## PPA OU : 3
## Glaucoma : 2
## - : 1
## ??glaucoma suspect but fields ok. See comments: 1
## 1 drusen for each eye : 1
## (Other) : 62
## UV.questionnaire_NIES.code UV.questionnaire_First.name Maiden.name
## Min. : 1.0 Mode:logical Mode:logical
## 1st Qu.:199.5 NA's:801 NA's:801
## Median :398.0
## Mean :398.3
## 3rd Qu.:598.5
## Max. :805.0
## NA's :10
## UV.questionnaire_Surname UV.questionnaire_Age UV.questionnaire_Sex
## Mode:logical Min. : 7.00 : 10
## NA's:801 1st Qu.:41.00 ,: 1
## Median :53.00 F:434
## Mean :52.39 M:356
## 3rd Qu.:65.00
## Max. :89.00
## NA's :13
## Lived.in.Norfolk.Island Location. From.age.in.years
## Min. : 0.00 :126 Min. : 0.000
## 1st Qu.:12.00 NZ :118 1st Qu.: 0.000
## Median :26.00 Sydney : 69 Median : 0.000
## Mean :27.52 AUS : 65 Mean : 8.367
## 3rd Qu.:40.00 NSW : 44 3rd Qu.:16.000
## Max. :89.00 QLD : 21 Max. :61.000
## NA's :32 (Other):358 NA's :131
## Duration.outside.Norfolk.Island Hair.colour Burns.Tans
## Min. : 0.00 Brown :493 burns then tans :462
## 1st Qu.:10.00 Black :122 never burns but tans:162
## Median :23.00 Blonde :115 burns not tan :110
## Mean :25.39 Red : 25 do not know : 24
## 3rd Qu.:39.00 NR : 18 : 18
## Max. :84.00 : 14 not applicable : 12
## NA's :112 (Other): 14 (Other) : 13
## Sunburn.and.pain Time.outside
## 2-10 times :412 : 17
## never :135 1/2 of the day :288
## once : 95 cannot judge : 47
## more than 10 times: 94 greater than 3/4 day:162
## do not know : 45 less than 1/4day :276
## : 15 none : 9
## (Other) : 5 u : 2
## Outdoors.and.hats Outdoors.and.sunglasses
## : 15 : 15
## 1/2 of the time:116 1/2 of the time: 85
## always :147 always :218
## can't judge : 14 can't judge : 11
## never :107 never :146
## seldom :168 seldom :161
## usually :234 usually :165
## Winter..Indoors.and.Outdoors At.school
## : 16 : 21
## 1/2 and 1/2 :424 do not know : 53
## do not know : 12 neither :376
## mostly indoors :186 yes, both :133
## mostly outdoors:163 yes, hat only :101
## yes, sunglasses only:117
##
## Feel.colder.than.people Arc.Welding If.yes..eye.protection
## : 16 : 18 :616
## 1/2 of the time: 82 don’t know: 23 don’t know: 8
## always : 32 No :562 No : 62
## cannot judge : 69 Yes :198 y : 2
## never :221 Yes :113
## seldom :280
## usually :101
## If.yes..flash.burn. Reason.for.sunglasses
## :615 glare :380
## don’t know: 9 :175
## No :126 driving : 75
## Yes : 51 protection from eye disease: 56
## medical requirement : 26
## fashion : 20
## (Other) : 69
## Do.not.wear.sunglasses X0531_niesid totaluvafmm
## :442 Min. : 1.0 Min. : 0.0
## Inconvenient : 98 1st Qu.:172.0 1st Qu.: 14.5
## Prescription glasses: 83 Median :368.0 Median : 28.1
## Not necessary : 72 Mean :384.6 Mean : 33.6
## Uncomfortable : 45 3rd Qu.:602.0 3rd Qu.: 47.8
## Decrease vision : 32 Max. :795.0 Max. :201.0
## (Other) : 29 NA's :152 NA's :160
## NIESMatch GLC_nies_code Glaucoma Diagnosis.GLC.known.previous.
## Mode:logical Min. : 40.0 :763 :763
## TRUE:801 1st Qu.:152.5 YES: 38 NEW DIAGNOSIS: 10
## Median :268.0 YES : 28
## Mean :291.6
## 3rd Qu.:378.5
## Max. :725.0
## NA's :763
## PIT GLC_griffith_code
## Min. :0.0000 :763
## 1st Qu.:0.0000 Â : 24
## Median :0.0000 1361059: 1
## Mean :0.4211 1361110: 1
## 3rd Qu.:1.0000 1361128: 1
## Max. :1.0000 1362450: 1
## NA's :763 (Other): 103. Subset data
quant.data_uvaf <- c("R.Logmar.VA", "RVA.with.PH", "L.Logmar.VA", "LVA.with.PH",
"R.Sph..pre.dilate.", "R.Cyl..pre.dilate.", "R.Axis..pre.dilate.", "L.Sph..pre.dilate.", "L.Cyl..pre.dilate.",
"L.Axis..pre.dilate.", "R.K.value.H", "R.K.Value.H.Axis", "R.K.value.V", "R.K.value.V.Axis", "L.K.value.H",
"L.K.value.H.Axis", "L.K.value.V", "L.K.value.V.Axis", "R.Pachimetry", "L.Pachimetry", "R.Axial.Length",
"L.Axial.Length", "R.AC.Depth", "L.AC.Depth", "R.IOP.mmHg", "L.IOP.mmHg", "R.CDR", "L.CDR", "totaluvafmm")
quant.data.subset_uvaf <- phen.data.uvaf[quant.data_uvaf]
head(quant.data.subset_uvaf)## R.Logmar.VA RVA.with.PH L.Logmar.VA LVA.with.PH R.Sph..pre.dilate.
## 1 0.02 0.02 -0.04 -0.04 0.25
## 2 0.10 0.10 0.16 0.10 0.00
## 3 0.00 0.00 0.00 0.00 1.25
## 4 0.30 0.10 0.08 0.08 1.25
## 5 0.00 0.00 -0.10 -0.10 1.25
## 6 0.24 0.16 0.36 0.14 4.00
## R.Cyl..pre.dilate. R.Axis..pre.dilate. L.Sph..pre.dilate.
## 1 0.00 0 0.25
## 2 -0.75 28 -0.50
## 3 -1.25 148 1.25
## 4 -0.25 97 1.50
## 5 -0.25 37 1.25
## 6 -0.50 46 3.75
## L.Cyl..pre.dilate. L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis
## 1 -0.50 79 42.00 2
## 2 -0.25 164 41.25 7
## 3 -0.25 24 NA NA
## 4 -0.50 81 44.75 5
## 5 -0.75 164 44.75 0
## 6 -0.75 180 42.00 8
## R.K.value.V R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## 1 43.00 92 42.50 5 43.50
## 2 42.25 97 41.50 168 42.00
## 3 NA NA NA NA NA
## 4 45.00 95 45.00 60 45.25
## 5 44.75 90 44.25 178 44.75
## 6 43.25 98 42.25 177 43.25
## L.K.value.V.Axis R.Pachimetry L.Pachimetry R.Axial.Length L.Axial.Length
## 1 95 532 554 24.31 24.10
## 2 78 608 612 25.02 25.21
## 3 NA 507 510 22.78 22.80
## 4 150 560 559 23.02 22.98
## 5 88 556 562 21.75 22.04
## 6 87 498 501 23.06 23.17
## R.AC.Depth L.AC.Depth R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR totaluvafmm
## 1 3.09 3.03 14 14 0.9 0.9 0.0
## 2 3.38 3.92 16 15 0.9 0.7 74.4
## 3 3.40 3.45 26 22 0.7 0.7 14.6
## 4 3.00 2.85 14 14 0.2 0.2 1.9
## 5 2.60 2.53 22 21 0.3 0.3 NA
## 6 2.94 3.04 18 20 0.6 0.6 1.04. Select adults only (>17 years)
phen.data.adults_uvaf<-phen.data.uvaf[phen.data.uvaf$Age.excel>17,]
quant.variables_uvaf<- c("R.Logmar.VA", "L.Logmar.VA",
"R.Sph..pre.dilate.", "R.Cyl..pre.dilate.", "R.Axis..pre.dilate.", "L.Sph..pre.dilate.", "L.Cyl..pre.dilate.",
"L.Axis..pre.dilate.", "R.K.value.H", "R.K.Value.H.Axis", "R.K.value.V", "R.K.value.V.Axis", "L.K.value.H",
"L.K.value.H.Axis", "L.K.value.V", "L.K.value.V.Axis", "R.Pachimetry", "L.Pachimetry", "R.Axial.Length",
"L.Axial.Length", "R.AC.Depth", "L.AC.Depth", "R.IOP.mmHg", "L.IOP.mmHg", "R.CDR", "L.CDR", "totaluvafmm")
quant.data.adults_uvaf<- phen.data.adults_uvaf[quant.variables_uvaf]
head(quant.data.adults_uvaf)## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate. R.Cyl..pre.dilate.
## 1 0.02 -0.04 0.25 0.00
## 2 0.10 0.16 0.00 -0.75
## 3 0.00 0.00 1.25 -1.25
## 4 0.30 0.08 1.25 -0.25
## 5 0.00 -0.10 1.25 -0.25
## 6 0.24 0.36 4.00 -0.50
## R.Axis..pre.dilate. L.Sph..pre.dilate. L.Cyl..pre.dilate.
## 1 0 0.25 -0.50
## 2 28 -0.50 -0.25
## 3 148 1.25 -0.25
## 4 97 1.50 -0.50
## 5 37 1.25 -0.75
## 6 46 3.75 -0.75
## L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis R.K.value.V
## 1 79 42.00 2 43.00
## 2 164 41.25 7 42.25
## 3 24 NA NA NA
## 4 81 44.75 5 45.00
## 5 164 44.75 0 44.75
## 6 180 42.00 8 43.25
## R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## 1 92 42.50 5 43.50
## 2 97 41.50 168 42.00
## 3 NA NA NA NA
## 4 95 45.00 60 45.25
## 5 90 44.25 178 44.75
## 6 98 42.25 177 43.25
## L.K.value.V.Axis R.Pachimetry L.Pachimetry R.Axial.Length L.Axial.Length
## 1 95 532 554 24.31 24.10
## 2 78 608 612 25.02 25.21
## 3 NA 507 510 22.78 22.80
## 4 150 560 559 23.02 22.98
## 5 88 556 562 21.75 22.04
## 6 87 498 501 23.06 23.17
## R.AC.Depth L.AC.Depth R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR totaluvafmm
## 1 3.09 3.03 14 14 0.9 0.9 0.0
## 2 3.38 3.92 16 15 0.9 0.7 74.4
## 3 3.40 3.45 26 22 0.7 0.7 14.6
## 4 3.00 2.85 14 14 0.2 0.2 1.9
## 5 2.60 2.53 22 21 0.3 0.3 NA
## 6 2.94 3.04 18 20 0.6 0.6 1.0summary(quant.data.adults_uvaf)## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate.
## Min. :-0.30000 Min. :-0.30000 Min. :-8.0000
## 1st Qu.:-0.08000 1st Qu.:-0.10000 1st Qu.: 0.0000
## Median : 0.02000 Median : 0.02000 Median : 0.5000
## Mean : 0.06608 Mean : 0.06359 Mean : 0.5455
## 3rd Qu.: 0.14000 3rd Qu.: 0.13000 3rd Qu.: 1.2500
## Max. : 2.00000 Max. : 1.70000 Max. : 8.5000
## NA's :3 NA's :6 NA's :10
## R.Cyl..pre.dilate. R.Axis..pre.dilate. L.Sph..pre.dilate.
## Min. :-11.0000 Min. : 0.00 Min. :-9.5000
## 1st Qu.: -0.7500 1st Qu.: 25.50 1st Qu.: 0.0000
## Median : -0.5000 Median : 85.00 Median : 0.5000
## Mean : -0.6497 Mean : 80.23 Mean : 0.5887
## 3rd Qu.: -0.2500 3rd Qu.:119.00 3rd Qu.: 1.2500
## Max. : 0.0000 Max. :180.00 Max. : 8.0000
## NA's :10 NA's :10 NA's :9
## L.Cyl..pre.dilate. L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis
## Min. :-9.7500 Min. : 0.00 Min. :36.00 Min. : 0.00
## 1st Qu.:-0.7500 1st Qu.: 13.00 1st Qu.:42.00 1st Qu.: 42.00
## Median :-0.5000 Median : 70.00 Median :43.00 Median : 91.00
## Mean :-0.6522 Mean : 72.25 Mean :42.97 Mean : 93.74
## 3rd Qu.:-0.2500 3rd Qu.:113.00 3rd Qu.:44.00 3rd Qu.:156.00
## Max. : 0.0000 Max. :180.00 Max. :48.25 Max. :180.00
## NA's :9 NA's :9 NA's :16 NA's :16
## R.K.value.V R.K.value.V.Axis L.K.value.H L.K.value.H.Axis
## Min. :37.00 Min. : 1.00 Min. :32.75 Min. : 0.0
## 1st Qu.:42.75 1st Qu.: 62.00 1st Qu.:42.00 1st Qu.: 24.0
## Median :43.75 Median : 89.00 Median :43.00 Median : 88.5
## Mean :43.80 Mean : 91.19 Mean :42.97 Mean : 87.8
## 3rd Qu.:45.00 3rd Qu.:127.00 3rd Qu.:44.00 3rd Qu.:147.0
## Max. :55.00 Max. :189.00 Max. :47.25 Max. :180.0
## NA's :16 NA's :16 NA's :15 NA's :15
## L.K.value.V L.K.value.V.Axis R.Pachimetry L.Pachimetry
## Min. :39.00 Min. : 0.00 Min. :428.0 Min. :424.0
## 1st Qu.:42.81 1st Qu.: 65.25 1st Qu.:527.0 1st Qu.:526.0
## Median :43.75 Median : 90.00 Median :546.0 Median :547.0
## Mean :43.87 Mean : 92.22 Mean :546.2 Mean :546.3
## 3rd Qu.:44.75 3rd Qu.:119.75 3rd Qu.:570.0 3rd Qu.:568.0
## Max. :52.00 Max. :180.00 Max. :656.0 Max. :658.0
## NA's :15 NA's :15 NA's :14 NA's :16
## R.Axial.Length L.Axial.Length R.AC.Depth L.AC.Depth
## Min. :20.95 Min. :21.29 Min. :2.090 Min. :2.000
## 1st Qu.:22.90 1st Qu.:22.87 1st Qu.:3.058 1st Qu.:3.040
## Median :23.50 Median :23.46 Median :3.310 Median :3.280
## Mean :23.56 Mean :23.54 Mean :3.320 Mean :3.306
## 3rd Qu.:24.09 3rd Qu.:24.11 3rd Qu.:3.550 3rd Qu.:3.553
## Max. :27.66 Max. :34.43 Max. :4.950 Max. :5.130
## NA's :13 NA's :13 NA's :13 NA's :13
## R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR
## Min. : 6.00 Min. : 8.00 Min. :0.0000 Min. :0.0000
## 1st Qu.:14.00 1st Qu.:14.00 1st Qu.:0.3000 1st Qu.:0.3000
## Median :16.00 Median :16.00 Median :0.4000 Median :0.4000
## Mean :15.88 Mean :16.06 Mean :0.4057 Mean :0.4022
## 3rd Qu.:18.00 3rd Qu.:18.00 3rd Qu.:0.5000 3rd Qu.:0.5000
## Max. :28.00 Max. :33.00 Max. :0.9900 Max. :0.9000
## NA's :2 NA's :3 NA's :19 NA's :17
## totaluvafmm
## Min. : 0.00
## 1st Qu.: 14.50
## Median : 28.00
## Mean : 33.53
## 3rd Qu.: 48.15
## Max. :201.00
## NA's :1385. Load and check ‘filtered’ data
ocular_data_uvaf <- read.csv('quant.data.adults_uvaf.csv', head = T, as.is = T, row.names = 1)
ocular_data_uvaf <- as.matrix(ocular_data_uvaf)
head(ocular_data_uvaf)## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate. R.Cyl..pre.dilate.
## 1 0.02 -0.04 0.25 0.00
## 2 0.10 0.16 0.00 -0.75
## 3 0.00 0.00 1.25 -1.25
## 4 0.30 0.08 1.25 -0.25
## 5 0.00 -0.10 1.25 -0.25
## 6 0.24 0.36 4.00 -0.50
## R.Axis..pre.dilate. L.Sph..pre.dilate. L.Cyl..pre.dilate.
## 1 0 0.25 -0.50
## 2 28 -0.50 -0.25
## 3 148 1.25 -0.25
## 4 97 1.50 -0.50
## 5 37 1.25 -0.75
## 6 46 3.75 -0.75
## L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis R.K.value.V
## 1 79 42.00 2 43.00
## 2 164 41.25 7 42.25
## 3 24 NA NA NA
## 4 81 44.75 5 45.00
## 5 164 44.75 0 44.75
## 6 180 42.00 8 43.25
## R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## 1 92 42.50 5 43.50
## 2 97 41.50 168 42.00
## 3 NA NA NA NA
## 4 95 45.00 60 45.25
## 5 90 44.25 178 44.75
## 6 98 42.25 177 43.25
## L.K.value.V.Axis R.Pachimetry L.Pachimetry R.Axial.Length L.Axial.Length
## 1 95 532 554 24.31 24.10
## 2 78 608 612 25.02 25.21
## 3 NA 507 510 22.78 22.80
## 4 150 560 559 23.02 22.98
## 5 88 556 562 21.75 22.04
## 6 87 498 501 23.06 23.17
## R.AC.Depth L.AC.Depth R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR totaluvafmm
## 1 3.09 3.03 14 14 0.9 0.9 0.0
## 2 3.38 3.92 16 15 0.9 0.7 74.4
## 3 3.40 3.45 26 22 0.7 0.7 14.6
## 4 3.00 2.85 14 14 0.2 0.2 1.9
## 5 2.60 2.53 22 21 0.3 0.3 NA
## 6 2.94 3.04 18 20 0.6 0.6 1.06. Perform data imputation
nbdim = estim_ncpPCA(ocular_data_uvaf, method = 'EM', method.cv="Kfold")##
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|=== | 5%
|
|==== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 12%
|
|========= | 13%
|
|========= | 14%
|
|========== | 15%
|
|=========== | 16%
|
|=========== | 17%
|
|============ | 18%
|
|============ | 19%
|
|============= | 20%
|
|============== | 21%
|
|============== | 22%
|
|=============== | 23%
|
|================ | 24%
|
|================ | 25%
|
|================= | 26%
|
|================== | 27%
|
|================== | 28%
|
|=================== | 29%
|
|==================== | 30%
|
|==================== | 31%
|
|===================== | 32%
|
|====================== | 33%
|
|====================== | 34%
|
|======================= | 35%
|
|======================== | 36%
|
|======================== | 37%
|
|========================= | 38%
|
|========================== | 39%
|
|========================== | 40%
|
|=========================== | 41%
|
|============================ | 42%
|
|============================ | 43%
|
|============================= | 44%
|
|============================== | 45%
|
|============================== | 46%
|
|=============================== | 47%
|
|================================ | 48%
|
|================================ | 49%
|
|================================= | 51%
|
|================================= | 52%
|
|================================== | 53%
|
|=================================== | 54%
|
|=================================== | 55%
|
|==================================== | 56%
|
|===================================== | 57%
|
|===================================== | 58%
|
|====================================== | 59%
|
|======================================= | 60%
|
|======================================= | 61%
|
|======================================== | 62%
|
|========================================= | 63%
|
|========================================= | 64%
|
|========================================== | 65%
|
|=========================================== | 66%
|
|=========================================== | 67%
|
|============================================ | 68%
|
|============================================= | 69%
|
|============================================= | 70%
|
|============================================== | 71%
|
|=============================================== | 72%
|
|=============================================== | 73%
|
|================================================ | 74%
|
|================================================= | 75%
|
|================================================= | 76%
|
|================================================== | 77%
|
|=================================================== | 78%
|
|=================================================== | 79%
|
|==================================================== | 80%
|
|===================================================== | 81%
|
|===================================================== | 82%
|
|====================================================== | 83%
|
|====================================================== | 84%
|
|======================================================= | 85%
|
|======================================================== | 86%
|
|======================================================== | 87%
|
|========================================================= | 88%
|
|========================================================== | 89%
|
|========================================================== | 90%
|
|=========================================================== | 91%
|
|============================================================ | 92%
|
|============================================================ | 93%
|
|============================================================= | 94%
|
|============================================================== | 95%
|
|============================================================== | 96%
|
|=============================================================== | 97%
|
|================================================================ | 98%
|
|================================================================ | 99%
|
|=================================================================| 100%nbdim## $ncp
## [1] 5
##
## $criterion
## 0 1 2 3 4 5
## 824625.1 826532.6 827067.7 827471.7 818691.8 798619.3res.comp = MIPCA(ocular_data_uvaf, ncp = nbdim$ncp, nboot = 1000)
png('indiv_plot_uvaf.png', width = 3200, height = 3200, res = 150)
plot.MIPCA(res.comp, choice = "ind.supp")
dev.off()## png
## 2png('var_plot_uvaf.png', width = 3200, height = 3200, res = 150)
plot.MIPCA(res.comp, choice = "var")
dev.off()## png
## 27. Write out imputed data frame and check summary statistics
imputed_phen_data_uvaf<-res.comp$res.imputePCA
head(imputed_phen_data_uvaf)## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate. R.Cyl..pre.dilate.
## 1 0.02 -0.04 0.25 0.00
## 2 0.10 0.16 0.00 -0.75
## 3 0.00 0.00 1.25 -1.25
## 4 0.30 0.08 1.25 -0.25
## 5 0.00 -0.10 1.25 -0.25
## 6 0.24 0.36 4.00 -0.50
## R.Axis..pre.dilate. L.Sph..pre.dilate. L.Cyl..pre.dilate.
## 1 0 0.25 -0.50
## 2 28 -0.50 -0.25
## 3 148 1.25 -0.25
## 4 97 1.50 -0.50
## 5 37 1.25 -0.75
## 6 46 3.75 -0.75
## L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis R.K.value.V
## 1 79 42.00000 2.0000 43.00000
## 2 164 41.25000 7.0000 42.25000
## 3 24 43.91437 145.9032 44.91517
## 4 81 44.75000 5.0000 45.00000
## 5 164 44.75000 0.0000 44.75000
## 6 180 42.00000 8.0000 43.25000
## R.K.value.V.Axis L.K.value.H L.K.value.H.Axis L.K.value.V
## 1 92.00000 42.50000 5.00000 43.50000
## 2 97.00000 41.50000 168.00000 42.00000
## 3 49.64518 44.06458 38.39706 44.88776
## 4 95.00000 45.00000 60.00000 45.25000
## 5 90.00000 44.25000 178.00000 44.75000
## 6 98.00000 42.25000 177.00000 43.25000
## L.K.value.V.Axis R.Pachimetry L.Pachimetry R.Axial.Length L.Axial.Length
## 1 95.0000 532 554 24.31 24.10
## 2 78.0000 608 612 25.02 25.21
## 3 137.2832 507 510 22.78 22.80
## 4 150.0000 560 559 23.02 22.98
## 5 88.0000 556 562 21.75 22.04
## 6 87.0000 498 501 23.06 23.17
## R.AC.Depth L.AC.Depth R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR totaluvafmm
## 1 3.09 3.03 14 14 0.9 0.9 0.00000
## 2 3.38 3.92 16 15 0.9 0.7 74.40000
## 3 3.40 3.45 26 22 0.7 0.7 14.60000
## 4 3.00 2.85 14 14 0.2 0.2 1.90000
## 5 2.60 2.53 22 21 0.3 0.3 23.98903
## 6 2.94 3.04 18 20 0.6 0.6 1.00000summary_stats_imputed_uvaf <-summary(imputed_phen_data_uvaf)
summary_stats_imputed_uvaf## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate.
## Min. :-0.30000 Min. :-0.30000 Min. :-8.0000
## 1st Qu.:-0.08000 1st Qu.:-0.10000 1st Qu.: 0.0000
## Median : 0.02000 Median : 0.02000 Median : 0.5000
## Mean : 0.06625 Mean : 0.06453 Mean : 0.5482
## 3rd Qu.: 0.14000 3rd Qu.: 0.14000 3rd Qu.: 1.2500
## Max. : 2.00000 Max. : 1.70000 Max. : 8.5000
## R.Cyl..pre.dilate. R.Axis..pre.dilate. L.Sph..pre.dilate.
## Min. :-11.0000 Min. : 0.00 Min. :-9.5000
## 1st Qu.: -0.7500 1st Qu.: 26.00 1st Qu.: 0.0000
## Median : -0.5000 Median : 85.00 Median : 0.5000
## Mean : -0.6536 Mean : 80.26 Mean : 0.5942
## 3rd Qu.: -0.2500 3rd Qu.:118.00 3rd Qu.: 1.2500
## Max. : 0.0000 Max. :180.00 Max. : 8.0000
## L.Cyl..pre.dilate. L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis
## Min. :-9.7500 Min. : 0.00 Min. :36.00 Min. : 0.00
## 1st Qu.:-0.7500 1st Qu.: 14.00 1st Qu.:42.00 1st Qu.: 44.00
## Median :-0.5000 Median : 71.00 Median :43.00 Median : 92.00
## Mean :-0.6564 Mean : 72.41 Mean :42.96 Mean : 93.87
## 3rd Qu.:-0.2500 3rd Qu.:112.00 3rd Qu.:44.00 3rd Qu.:154.00
## Max. : 0.0000 Max. :180.00 Max. :48.25 Max. :180.00
## R.K.value.V R.K.value.V.Axis L.K.value.H L.K.value.H.Axis
## Min. :37.00 Min. : 1.00 Min. :32.75 Min. : 0.00
## 1st Qu.:42.75 1st Qu.: 63.00 1st Qu.:42.00 1st Qu.: 26.00
## Median :43.75 Median : 89.00 Median :43.00 Median : 88.00
## Mean :43.79 Mean : 91.07 Mean :42.96 Mean : 87.74
## 3rd Qu.:44.86 3rd Qu.:124.00 3rd Qu.:44.00 3rd Qu.:145.00
## Max. :55.00 Max. :189.00 Max. :47.25 Max. :180.00
## L.K.value.V L.K.value.V.Axis R.Pachimetry L.Pachimetry
## Min. :39.00 Min. : 0.00 Min. :428.0 Min. :424.0
## 1st Qu.:42.88 1st Qu.: 68.00 1st Qu.:526.3 1st Qu.:526.0
## Median :43.75 Median : 90.00 Median :546.0 Median :547.0
## Mean :43.87 Mean : 92.32 Mean :546.0 Mean :546.1
## 3rd Qu.:44.75 3rd Qu.:119.00 3rd Qu.:570.0 3rd Qu.:568.0
## Max. :52.00 Max. :180.00 Max. :656.0 Max. :658.0
## R.Axial.Length L.Axial.Length R.AC.Depth L.AC.Depth
## Min. :20.95 Min. :21.29 Min. :2.090 Min. :2.000
## 1st Qu.:22.91 1st Qu.:22.87 1st Qu.:3.060 1st Qu.:3.040
## Median :23.51 Median :23.47 Median :3.310 Median :3.290
## Mean :23.56 Mean :23.54 Mean :3.322 Mean :3.308
## 3rd Qu.:24.10 3rd Qu.:24.11 3rd Qu.:3.560 3rd Qu.:3.560
## Max. :27.66 Max. :34.43 Max. :4.950 Max. :5.130
## R.IOP.mmHg L.IOP.mmHg R.CDR L.CDR
## Min. : 6.00 Min. : 8.00 Min. :0.0000 Min. :0.0000
## 1st Qu.:14.00 1st Qu.:14.00 1st Qu.:0.3000 1st Qu.:0.3000
## Median :16.00 Median :16.00 Median :0.4000 Median :0.4000
## Mean :15.88 Mean :16.06 Mean :0.4076 Mean :0.4038
## 3rd Qu.:18.00 3rd Qu.:18.00 3rd Qu.:0.5000 3rd Qu.:0.5000
## Max. :28.00 Max. :33.00 Max. :0.9900 Max. :0.9000
## totaluvafmm
## Min. : 0.00
## 1st Qu.: 16.70
## Median : 29.60
## Mean : 33.46
## 3rd Qu.: 44.40
## Max. :201.008. Data cleaning
imputed_phen_data_uvaf %>%
as.data.frame() %>%
keep(is.numeric) %>%
gather() %>%
ggplot(aes(value)) + facet_wrap(~ key, scales = "free") + geom_histogram(fill = 'grey', colour = "black")## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.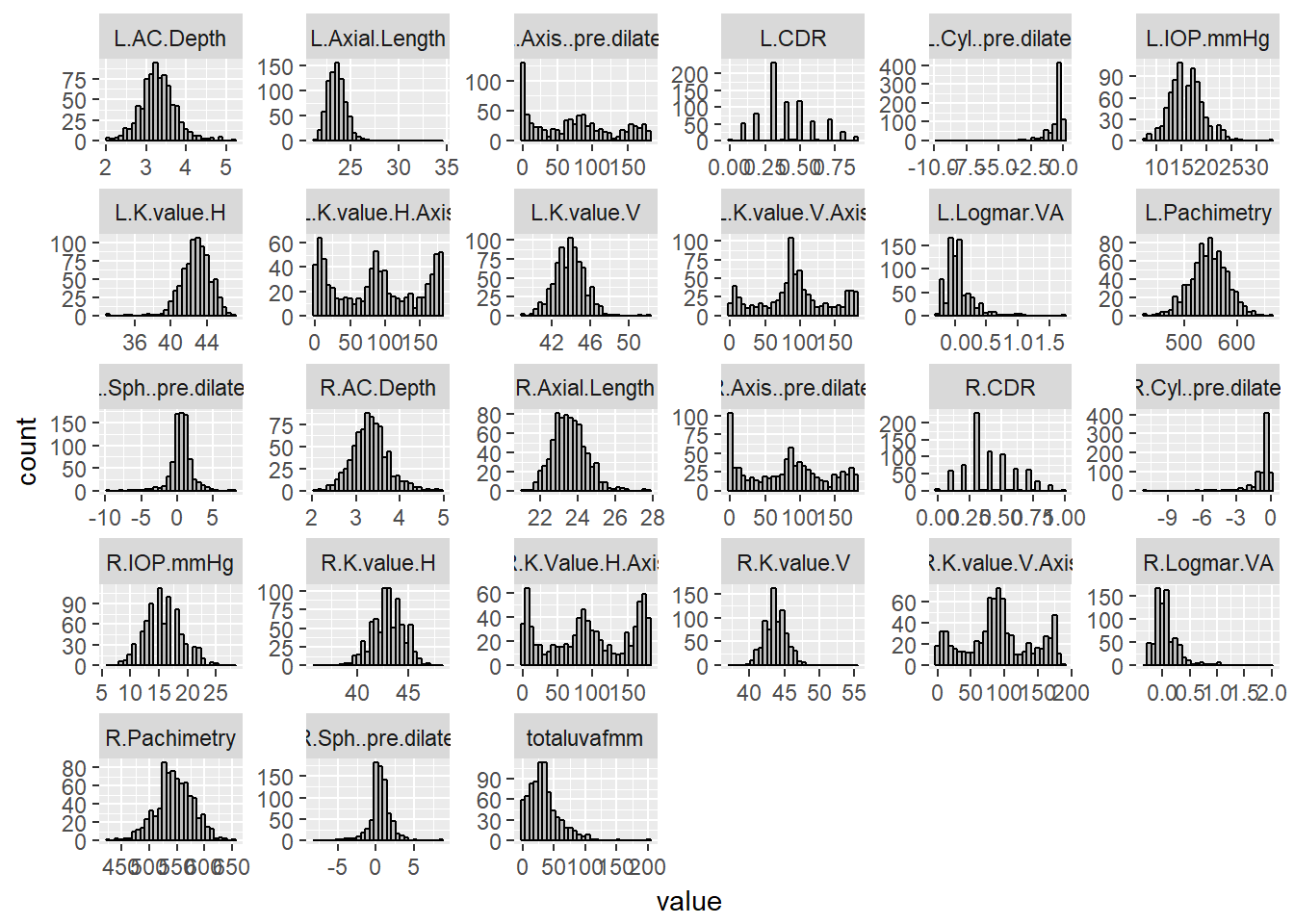
9. QC and summary statistics
skewness_imp_uvaf<-skewness(imputed_phen_data_uvaf)
kurtosis_imp_uvaf<-kurtosis(imputed_phen_data_uvaf)
# summary stats
summary_stats_imputed_uvaf<-rbind(summary_stats_imputed_uvaf, skewness_imp_uvaf, kurtosis_imp_uvaf)
summary_stats_imputed_uvaf## R.Logmar.VA L.Logmar.VA
## "Min. :-0.30000 " "Min. :-0.30000 "
## "1st Qu.:-0.08000 " "1st Qu.:-0.10000 "
## "Median : 0.02000 " "Median : 0.02000 "
## "Mean : 0.06625 " "Mean : 0.06453 "
## "3rd Qu.: 0.14000 " "3rd Qu.: 0.14000 "
## "Max. : 2.00000 " "Max. : 1.70000 "
## skewness_imp_uvaf "2.59211089274108" "1.98559157007324"
## kurtosis_imp_uvaf "15.6538434136427" "9.79306589470692"
## R.Sph..pre.dilate. R.Cyl..pre.dilate.
## "Min. :-8.0000 " "Min. :-11.0000 "
## "1st Qu.: 0.0000 " "1st Qu.: -0.7500 "
## "Median : 0.5000 " "Median : -0.5000 "
## "Mean : 0.5482 " "Mean : -0.6536 "
## "3rd Qu.: 1.2500 " "3rd Qu.: -0.2500 "
## "Max. : 8.5000 " "Max. : 0.0000 "
## skewness_imp_uvaf "-0.497785148561143" "-5.06920609089807"
## kurtosis_imp_uvaf "9.82756542236217" "50.4664156747398"
## R.Axis..pre.dilate. L.Sph..pre.dilate.
## "Min. : 0.00 " "Min. :-9.5000 "
## "1st Qu.: 26.00 " "1st Qu.: 0.0000 "
## "Median : 85.00 " "Median : 0.5000 "
## "Mean : 80.26 " "Mean : 0.5942 "
## "3rd Qu.:118.00 " "3rd Qu.: 1.2500 "
## "Max. :180.00 " "Max. : 8.0000 "
## skewness_imp_uvaf "0.067109758301877" "-0.703446131408983"
## kurtosis_imp_uvaf "1.90102600391832" "10.4651241460371"
## L.Cyl..pre.dilate. L.Axis..pre.dilate.
## "Min. :-9.7500 " "Min. : 0.00 "
## "1st Qu.:-0.7500 " "1st Qu.: 14.00 "
## "Median :-0.5000 " "Median : 71.00 "
## "Mean :-0.6564 " "Mean : 72.41 "
## "3rd Qu.:-0.2500 " "3rd Qu.:112.00 "
## "Max. : 0.0000 " "Max. :180.00 "
## skewness_imp_uvaf "-4.73814034449544" "0.299770145841937"
## kurtosis_imp_uvaf "35.5986818044468" "1.88926703932912"
## R.K.value.H R.K.Value.H.Axis
## "Min. :36.00 " "Min. : 0.00 "
## "1st Qu.:42.00 " "1st Qu.: 44.00 "
## "Median :43.00 " "Median : 92.00 "
## "Mean :42.96 " "Mean : 93.87 "
## "3rd Qu.:44.00 " "3rd Qu.:154.00 "
## "Max. :48.25 " "Max. :180.00 "
## skewness_imp_uvaf "-0.32799389328225" "-0.0875824503464911"
## kurtosis_imp_uvaf "3.66595676488688" "1.7299694740147"
## R.K.value.V R.K.value.V.Axis
## "Min. :37.00 " "Min. : 1.00 "
## "1st Qu.:42.75 " "1st Qu.: 63.00 "
## "Median :43.75 " "Median : 89.00 "
## "Mean :43.79 " "Mean : 91.07 "
## "3rd Qu.:44.86 " "3rd Qu.:124.00 "
## "Max. :55.00 " "Max. :189.00 "
## skewness_imp_uvaf "0.434159616427538" "0.0478589094219236"
## kurtosis_imp_uvaf "6.27946856444013" "2.29283887011322"
## L.K.value.H L.K.value.H.Axis
## "Min. :32.75 " "Min. : 0.00 "
## "1st Qu.:42.00 " "1st Qu.: 26.00 "
## "Median :43.00 " "Median : 88.00 "
## "Mean :42.96 " "Mean : 87.74 "
## "3rd Qu.:44.00 " "3rd Qu.:145.00 "
## "Max. :47.25 " "Max. :180.00 "
## skewness_imp_uvaf "-0.936745093906448" "0.0571539383782265"
## kurtosis_imp_uvaf "6.94859925843675" "1.69523308431538"
## L.K.value.V L.K.value.V.Axis
## "Min. :39.00 " "Min. : 0.00 "
## "1st Qu.:42.88 " "1st Qu.: 68.00 "
## "Median :43.75 " "Median : 90.00 "
## "Mean :43.87 " "Mean : 92.32 "
## "3rd Qu.:44.75 " "3rd Qu.:119.00 "
## "Max. :52.00 " "Max. :180.00 "
## skewness_imp_uvaf "0.232280803959146" "-0.0268648206524532"
## kurtosis_imp_uvaf "4.1057499370676" "2.38730261399806"
## R.Pachimetry L.Pachimetry
## "Min. :428.0 " "Min. :424.0 "
## "1st Qu.:526.3 " "1st Qu.:526.0 "
## "Median :546.0 " "Median :547.0 "
## "Mean :546.0 " "Mean :546.1 "
## "3rd Qu.:570.0 " "3rd Qu.:568.0 "
## "Max. :656.0 " "Max. :658.0 "
## skewness_imp_uvaf "-0.130181733965154" "-0.0960875038390799"
## kurtosis_imp_uvaf "3.11241295669933" "3.244660608979"
## R.Axial.Length L.Axial.Length
## "Min. :20.95 " "Min. :21.29 "
## "1st Qu.:22.91 " "1st Qu.:22.87 "
## "Median :23.51 " "Median :23.47 "
## "Mean :23.56 " "Mean :23.54 "
## "3rd Qu.:24.10 " "3rd Qu.:24.11 "
## "Max. :27.66 " "Max. :34.43 "
## skewness_imp_uvaf "0.623763085513932" "2.03373094331034"
## kurtosis_imp_uvaf "4.20615470168677" "20.6567130030489"
## R.AC.Depth L.AC.Depth
## "Min. :2.090 " "Min. :2.000 "
## "1st Qu.:3.060 " "1st Qu.:3.040 "
## "Median :3.310 " "Median :3.290 "
## "Mean :3.322 " "Mean :3.308 "
## "3rd Qu.:3.560 " "3rd Qu.:3.560 "
## "Max. :4.950 " "Max. :5.130 "
## skewness_imp_uvaf "0.384887959156102" "0.411011834939057"
## kurtosis_imp_uvaf "3.99884365766774" "4.26971992614078"
## R.IOP.mmHg L.IOP.mmHg
## "Min. : 6.00 " "Min. : 8.00 "
## "1st Qu.:14.00 " "1st Qu.:14.00 "
## "Median :16.00 " "Median :16.00 "
## "Mean :15.88 " "Mean :16.06 "
## "3rd Qu.:18.00 " "3rd Qu.:18.00 "
## "Max. :28.00 " "Max. :33.00 "
## skewness_imp_uvaf "0.218520840080988" "0.456103295787158"
## kurtosis_imp_uvaf "3.09145271634485" "3.91169191866095"
## R.CDR L.CDR
## "Min. :0.0000 " "Min. :0.0000 "
## "1st Qu.:0.3000 " "1st Qu.:0.3000 "
## "Median :0.4000 " "Median :0.4000 "
## "Mean :0.4076 " "Mean :0.4038 "
## "3rd Qu.:0.5000 " "3rd Qu.:0.5000 "
## "Max. :0.9900 " "Max. :0.9000 "
## skewness_imp_uvaf "0.526922774463583" "0.509778315846961"
## kurtosis_imp_uvaf "2.74290904762968" "2.74829620989337"
## totaluvafmm
## "Min. : 0.00 "
## "1st Qu.: 16.70 "
## "Median : 29.60 "
## "Mean : 33.46 "
## "3rd Qu.: 44.40 "
## "Max. :201.00 "
## skewness_imp_uvaf "1.36499925675976"
## kurtosis_imp_uvaf "6.81904459922986"10. Correlation analysis
pearson_imputed_data_uvaf<-rcorr(imputed_phen_data_uvaf, type="pearson")
#extract correlation coefficients
pearson_coeff_uvaf<-rcorr(imputed_phen_data_uvaf, type="pearson")$r
#extract p-values for correlations
pearson_pval_uvaf<-rcorr(imputed_phen_data_uvaf, type="pearson")$P
#bind coefficients and p-values
pearson_phen_data_uvaf<-rbind(pearson_coeff_uvaf, pearson_pval_uvaf)
head(pearson_phen_data_uvaf)## R.Logmar.VA L.Logmar.VA R.Sph..pre.dilate.
## R.Logmar.VA 1.00000000 0.44554620 0.01740867
## L.Logmar.VA 0.44554620 1.00000000 0.05500151
## R.Sph..pre.dilate. 0.01740867 0.05500151 1.00000000
## R.Cyl..pre.dilate. -0.36890659 -0.25211155 -0.03366801
## R.Axis..pre.dilate. 0.13854742 0.09972247 0.05003542
## L.Sph..pre.dilate. 0.03409520 0.04629321 0.73029352
## R.Cyl..pre.dilate. R.Axis..pre.dilate.
## R.Logmar.VA -0.36890659 0.13854742
## L.Logmar.VA -0.25211155 0.09972247
## R.Sph..pre.dilate. -0.03366801 0.05003542
## R.Cyl..pre.dilate. 1.00000000 -0.16925988
## R.Axis..pre.dilate. -0.16925988 1.00000000
## L.Sph..pre.dilate. -0.02703691 0.06864150
## L.Sph..pre.dilate. L.Cyl..pre.dilate.
## R.Logmar.VA 0.03409520 -0.215348171
## L.Logmar.VA 0.04629321 -0.335474654
## R.Sph..pre.dilate. 0.73029352 0.005414533
## R.Cyl..pre.dilate. -0.02703691 0.597233744
## R.Axis..pre.dilate. 0.06864150 -0.073540295
## L.Sph..pre.dilate. 1.00000000 -0.116741842
## L.Axis..pre.dilate. R.K.value.H R.K.Value.H.Axis
## R.Logmar.VA 0.05281465 -0.01392821 -0.018950297
## L.Logmar.VA 0.11239630 -0.03285066 -0.015110307
## R.Sph..pre.dilate. 0.03744512 -0.10744079 -0.049268956
## R.Cyl..pre.dilate. -0.07210691 0.08760364 0.017104717
## R.Axis..pre.dilate. -0.14258283 0.02183027 0.229646327
## L.Sph..pre.dilate. 0.05226560 -0.08368679 -0.008651087
## R.K.value.V R.K.value.V.Axis L.K.value.H
## R.Logmar.VA 0.08855648 0.012347435 -0.01000858
## L.Logmar.VA 0.03052644 0.008232785 -0.08934292
## R.Sph..pre.dilate. -0.06543496 0.060919398 -0.09981024
## R.Cyl..pre.dilate. -0.25808574 -0.011089058 0.07809858
## R.Axis..pre.dilate. 0.04771139 -0.191418829 0.03891460
## L.Sph..pre.dilate. -0.05540387 0.040524615 -0.15266391
## L.K.value.H.Axis L.K.value.V L.K.value.V.Axis
## R.Logmar.VA 0.03670637 0.07628865 -0.054508676
## L.Logmar.VA 0.09330733 0.07414806 -0.029672619
## R.Sph..pre.dilate. 0.08122655 -0.08328343 -0.032530066
## R.Cyl..pre.dilate. -0.01938538 -0.21427740 -0.003382738
## R.Axis..pre.dilate. -0.10884355 0.04783105 0.145976074
## L.Sph..pre.dilate. 0.05134335 -0.08285544 -0.021367228
## R.Pachimetry L.Pachimetry R.Axial.Length
## R.Logmar.VA -0.07127554 -0.052555626 -0.005651912
## L.Logmar.VA -0.07029725 -0.046377740 0.031847893
## R.Sph..pre.dilate. -0.02793746 -0.022103894 -0.482121780
## R.Cyl..pre.dilate. 0.06306713 0.062583717 -0.080955086
## R.Axis..pre.dilate. -0.03752342 -0.032809722 -0.036383894
## L.Sph..pre.dilate. -0.02193828 -0.009068986 -0.463226389
## L.Axial.Length R.AC.Depth L.AC.Depth R.IOP.mmHg
## R.Logmar.VA -0.01415605 -0.03341014 -0.02881020 -0.025892981
## L.Logmar.VA 0.03694522 -0.03143276 -0.05265482 -0.015281245
## R.Sph..pre.dilate. -0.39590302 -0.36071477 -0.34336245 -0.049302805
## R.Cyl..pre.dilate. -0.09784082 -0.08724048 -0.06086049 0.043557229
## R.Axis..pre.dilate. -0.06516755 -0.01208040 -0.01773704 0.008059977
## L.Sph..pre.dilate. -0.44973673 -0.32534938 -0.29171067 -0.073548376
## L.IOP.mmHg R.CDR L.CDR totaluvafmm
## R.Logmar.VA -0.027237285 0.17646064 0.11520935 -0.1422378286
## L.Logmar.VA 0.026534608 0.19112044 0.17303603 -0.1018966500
## R.Sph..pre.dilate. -0.047476695 0.06372020 0.07338004 -0.0011464564
## R.Cyl..pre.dilate. 0.029462272 -0.13574853 -0.10606450 0.1511516008
## R.Axis..pre.dilate. -0.007708397 0.05572869 0.06561538 -0.0004132163
## L.Sph..pre.dilate. -0.078120952 0.08532067 0.09537028 0.031669603511. Plotting
# correlation plot
title<- "Pearson correlation of imputed phenotypic data with UVAF"
corrplot.mixed(pearson_coeff_uvaf, p.mat=pearson_pval_uvaf, sig.level = 0.05, insig="blank", tl.pos = 'lt',
tl.col="black", tl.cex=0.60, mar=c(0,0,1,0), number.cex = 0.50, title=title)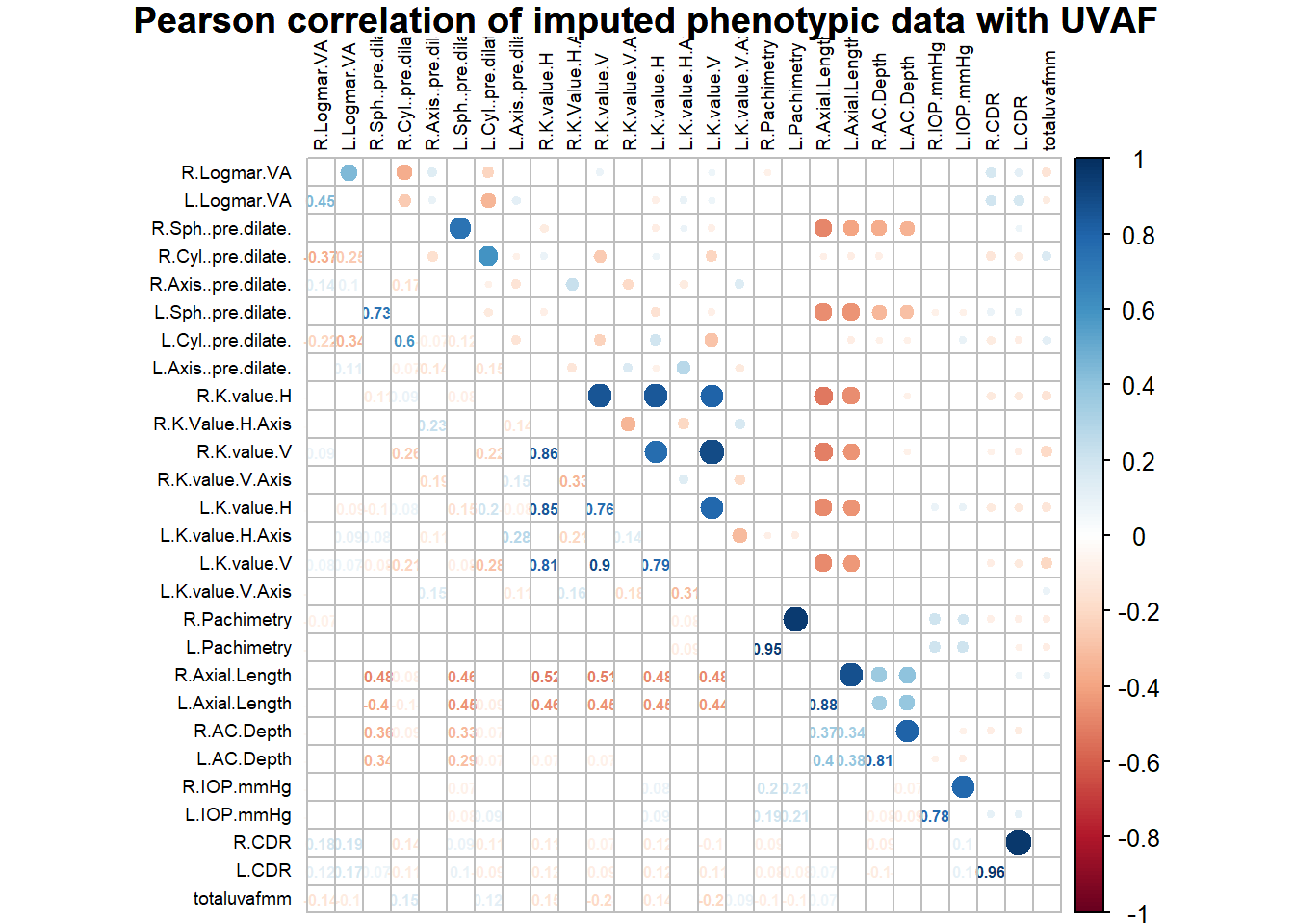
Discussion
The results of this multiple PCA imputation provides me with a complete data set, including the conjunctival UVAF variable. From the variable factor plot, it appears that the spread for the UVAF is larger than all other variables, which was expected since it had the most number of missing values. Unlike the logMAR with PH variables, this uncertainty is not large enough to exclude the UVAF variable, so it will be included for subsequent analysis. From the histograms of the imputed data, the axes variables evidently have trimodal distributions. It is unknown whether this type of distribution would affect the results of the PCA. I will run a PCA with all of the variables, regardless. All correlations shown in the correlation plot are statistically significant (p < 0.05). The Pearson correlation plots show several strongly positively correlated variables, which are the left and right measurements of the same variable and is expected. There are also some negatively correlated variables, suggesting that the Sph pre-dilate variables are correlated with axial length and anterior chamber depth. Clinically, these correlations make sense since the Sph pre-dilate values measure astigmatism, and unusual axial length and ACD contribute to it. It also shows that keratometry measurements are negatively correlated with axial length. This is also logically sound because keratometry tests measure corneal curvature and axial length directly affects it. The plot also shows that axial length and anterior chamber depth are positively correlated.