| tags: [ Data Cleaning Imputation PCA R ] categories: [Experiments Coding ]
Example of data imputation with missMDA
Introduction
This entry will show the results of a single imputation using the missMDA package. The missMDA package imputes quantitative variables using principal component analysis (PCA).
Methods and Results
- load data
phen.data.age <- read.csv('C:/Users/Martha/Documents/Honours/Project/honours.project/Data/NIES_master_database-age.csv')
phen.data.adults<-phen.data.age[phen.data.age$Age.excel>17,]
quant.variables<- c("R.K.value.H", "R.K.Value.H.Axis", "R.K.value.V", "R.K.value.V.Axis", "L.K.value.H",
"L.K.value.H.Axis", "L.K.value.V", "L.K.value.V.Axis", "R.Pachimetry", "L.Pachimetry", "R.Axial.Length",
"L.Axial.Length", "AC.Depth.R", "AC.Depth.L", "R.IOP.mmHg", "L.IOP.mmHg", "CDR.RE", "CDR.LE")
quant.data.adults<- phen.data.adults[quant.variables]- install relevant packages
install.packages('missMDA', dependencies = TRUE, repos = "http://cran.us.r-project.org")## Installing package into 'C:/Users/Martha/Documents/R/win-library/3.4'
## (as 'lib' is unspecified)## package 'missMDA' successfully unpacked and MD5 sums checked
##
## The downloaded binary packages are in
## C:\Users\Martha\AppData\Local\Temp\RtmpaopRRl\downloaded_packagesrequire(missMDA)## Loading required package: missMDArequire(FactoMineR)## Loading required package: FactoMineR- Estimate the number of dimensions to be used for the reconstruction formula
data(quant.data.adults)## Warning in data(quant.data.adults): data set 'quant.data.adults' not foundnb = estim_ncpPCA(quant.data.adults)
nb## $ncp
## [1] 0
##
## $criterion
## 0 1 2 3 4 5
## 805.7256 908.9445 966.5074 1029.5131 1088.5948 880.7742- Impute the data with the number of dimensions previously calculated
res.comp = imputePCA(quant.data.adults,ncp=nb$ncp)
head(res.comp$completeObs) #view imputed dataset## R.K.value.H R.K.Value.H.Axis R.K.value.V R.K.value.V.Axis L.K.value.H
## 1 42.00000 2.00000 43.00000 92.00000 42.50000
## 2 41.25000 7.00000 42.25000 97.00000 41.50000
## 3 42.96687 93.74108 43.79634 91.18098 42.96871
## 4 44.75000 5.00000 45.00000 95.00000 45.00000
## 5 44.75000 0.00000 44.75000 90.00000 44.25000
## 6 42.00000 8.00000 43.25000 98.00000 42.25000
## L.K.value.H.Axis L.K.value.V L.K.value.V.Axis R.Pachimetry L.Pachimetry
## 1 5.00000 43.50000 95.00000 532 554
## 2 168.00000 42.00000 78.00000 608 612
## 3 87.79947 43.87197 92.21768 507 510
## 4 60.00000 45.25000 150.00000 560 559
## 5 178.00000 44.75000 88.00000 556 562
## 6 177.00000 43.25000 87.00000 498 501
## R.Axial.Length L.Axial.Length AC.Depth.R AC.Depth.L R.IOP.mmHg
## 1 24.31 24.10 3.09 3.03 14
## 2 25.02 25.21 3.38 3.92 16
## 3 22.78 22.80 3.40 3.45 26
## 4 23.02 22.98 3.00 2.85 14
## 5 21.75 22.04 2.60 2.53 22
## 6 23.06 23.17 2.94 3.04 18
## L.IOP.mmHg CDR.RE CDR.LE
## 1 14 0.9 0.9
## 2 15 0.9 0.7
## 3 22 0.7 0.7
## 4 14 0.2 0.2
## 5 21 0.3 0.3
## 6 20 0.6 0.6- Perform PCA on the imputed data set and plot uncertainties
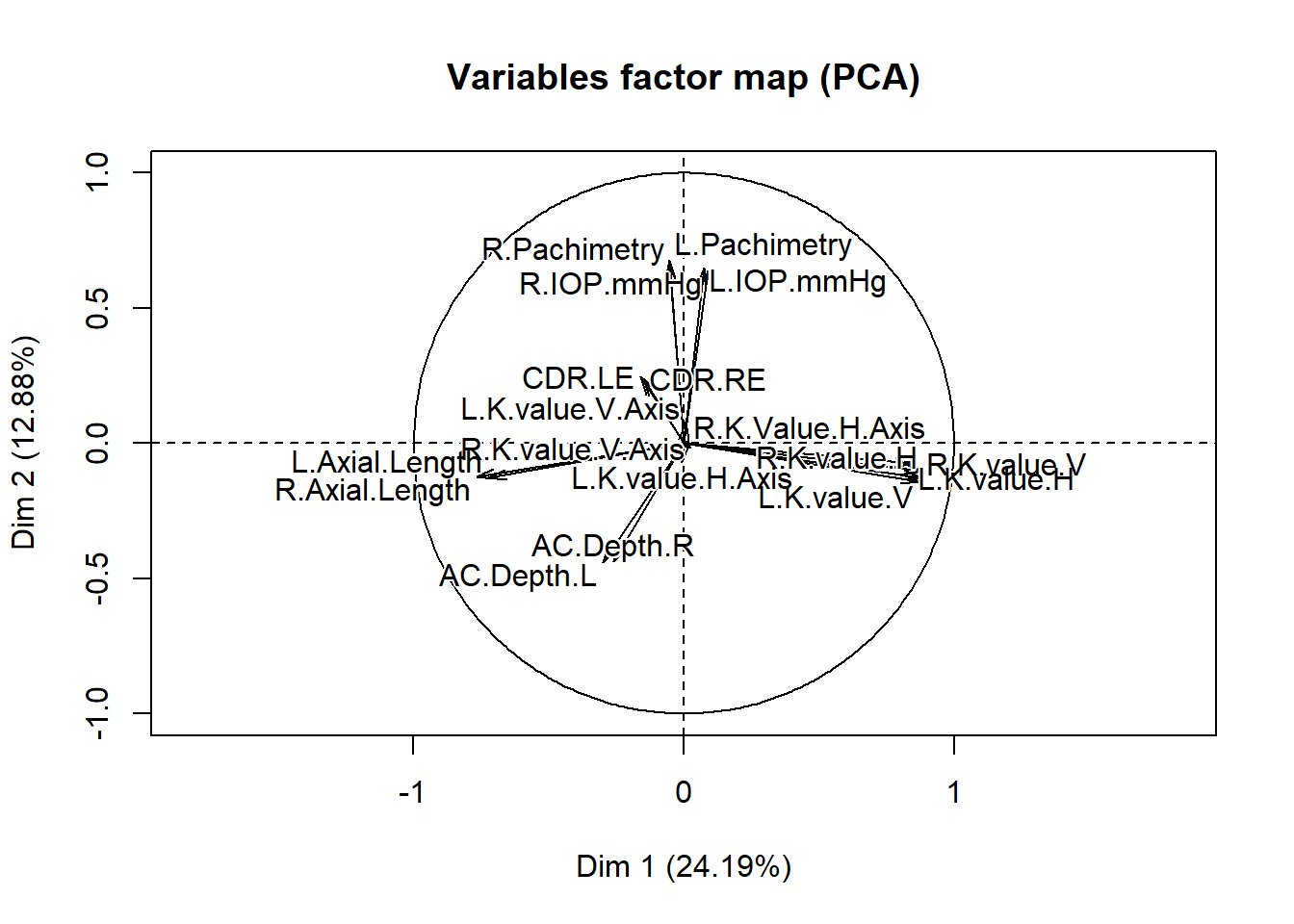
res.pca = PCA(res.comp$completeObs)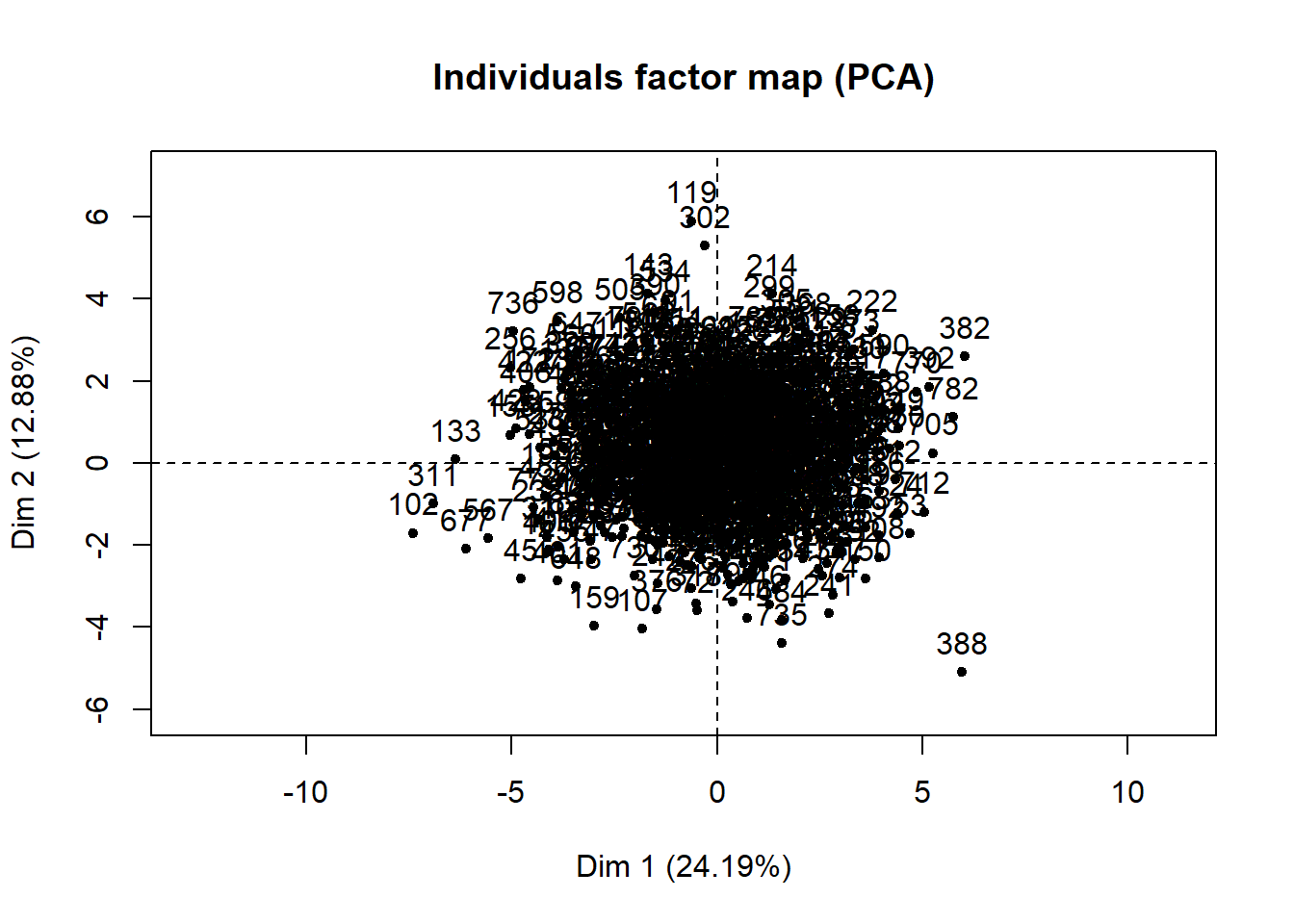
- Perform multiple imputations
“res.comp = MIPCA(quant.data.adults, ncp = nb$ncp, nboot = 1000)”
Discussion
The single imputation has produced a complete dataset. It is difficult to interpret the individuals factor map in detail because there are a large number of individuals in the study, and each dot represents an individual. If any of the participants had a signifiant amount of data missing, a circle around the dot would be present signifying the uncertainty of the imputed values. The larger the area of the circle, the higher the uncertainty. However, there does not appear to be any that are clearly visible in this plot, which suggests that the imputed values for each individual fits well. It also possible that any circles that are present are not visible because of the dense cluster of dots in the plot. The variables factor map shows the uncertainty of the imputed values for each variable.
https://cran.r-project.org/web/packages/missMDA/missMDA.pdf http://factominer.free.fr/missMDA/PCA.html http://www.statpower.net/Content/312/R%20Stuff/PCA.html
The results of this PCA imputation does not show the variation for each individual or variable as it was only performed once. Variation, or uncertainty, of the imputed values can be determined by performing multiple imputations.