| tags: [ Gaston GCTA Genomic Data GWAS Heritability MLMA ] categories: [Experiments Coding ]
SNP heritability
Introduction
SNP heritability of index SNPs are non-sense. The numbers I am obtaining are either too high or too low. Therefore, I want to compare GWAS results using the lmm/LRT method vs lm/Wald method. The wald test will also generate beta-values which are alternative values that indicate the effect size of SNVs.
Methods and Results
require(magrittr)
require(dplyr)
require(gaston)
require(qqman)Load relevant data
nies_heritable_pheno <- read.csv('C:/Users/Martha/Documents/Honours/Project/honours.project/Data/nies_heritable_pheno.csv', header = T)
merged_nies_210818 <- read.bed.matrix("C:/Users/Martha/Documents/Honours/Project/honours.project/Data/merged_nies/merged_nies_geno_210818.bed")## Reading C:/Users/Martha/Documents/Honours/Project/honours.project/Data/merged_nies/merged_nies_geno_210818.fam
## Reading C:/Users/Martha/Documents/Honours/Project/honours.project/Data/merged_nies/merged_nies_geno_210818.bim
## Reading C:/Users/Martha/Documents/Honours/Project/honours.project/Data/merged_nies/merged_nies_geno_210818.bed
## ped stats and snps stats have been set.
## 'p' has been set.
## 'mu' and 'sigma' have been set.merged_nies_GRM <- GRM(merged_nies_210818)
merged_nies_eiK <- eigen(merged_nies_GRM)
merged_nies_eiK$values[ merged_nies_eiK$values < 0] <- 0
merged_nies_PC <- sweep(merged_nies_eiK$vectors, 2, sqrt(merged_nies_eiK$values), "*")1. GWAS using lmm and lrt
l_KvalH_gwas <- association.test(merged_nies_210818, nies_heritable_pheno$L.K.value.H, method="lmm", test = "lrt", response = "quantitative", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
l_KvalH_gwas <- na.omit(l_KvalH_gwas)
l_kvalH_filtered <- l_KvalH_gwas %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
l_kvalH_meff <- l_kvalH_filtered %>% filter(p < 1.84e-7)
l_kvalH_sig <- do.call(rbind, lapply(split(l_kvalH_meff,l_kvalH_meff$chr), function(x) {return(x[which.min(x$p),])}))
l_kvalH_res <- NULL
for (i in l_kvalH_sig$id) {
snpID <- i
snpCHR <- l_kvalH_filtered[l_kvalH_filtered$id == snpID,]$chr
snpPOS <- l_kvalH_filtered[l_kvalH_filtered$id ==snpID,]$pos
sig.peak <- l_kvalH_filtered %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
l_kvalH_res <- rbind(l_kvalH_res, sig.peak)
}
l_kvalH_res## chr pos id A1 A2 freqA2 h2 LRT
## 1 2 32321350 rs148319856 C A 0.9826087 0.22244002 33.29515
## 2 2 32367072 rs115270434 C A 0.9855072 0.17567169 20.65630
## 3 3 77739700 rs1163757 A G 0.9869565 0.03021585 32.17915
## 4 6 82442490 rs114928787 T C 0.9855072 0.00000000 43.16564
## 5 6 82442833 rs116172930 C A 0.9855072 0.00000000 43.16564
## 6 6 82443802 rs74436554 C T 0.9855072 0.00000000 43.16564
## 7 6 82444791 rs78056249 C G 0.9855072 0.00000000 43.16564
## 8 6 82445716 rs77941466 C T 0.9855072 0.00000000 43.16564
## 9 6 82447004 rs116461493 T C 0.9855072 0.00000000 43.16564
## 10 6 82447732 rs77666578 G A 0.9855072 0.00000000 43.16564
## 11 6 82451563 rs75189229 A G 0.9855072 0.00000000 43.16564
## 12 6 82453938 rs117178778 A G 0.9855072 0.00000000 43.16564
## 13 6 82454058 rs116246536 T G 0.9855072 0.00000000 43.16564
## 14 6 82454140 rs78427866 A G 0.9855072 0.00000000 43.16564
## 15 6 82454380 rs147257041 A G 0.9855072 0.00000000 43.16564
## 16 6 82454449 rs140711073 T C 0.9855072 0.00000000 43.16564
## 17 6 82454505 rs142248448 T C 0.9855072 0.00000000 43.16564
## 18 6 82454620 rs114442506 A G 0.9855072 0.00000000 43.16564
## 19 6 82456537 rs145558293 A G 0.9855072 0.00000000 43.16564
## 20 6 82458393 rs77391407 C T 0.9855072 0.00000000 43.16564
## 21 6 82458546 rs77103853 G C 0.9855072 0.00000000 43.16564
## 22 6 82459011 rs115096710 T C 0.9855072 0.00000000 43.16564
## 23 6 82459713 rs77245672 G C 0.9855072 0.00000000 43.16564
## 24 6 82459810 rs77599496 T A 0.9855072 0.00000000 43.16564
## 25 6 82459890 rs77598967 G A 0.9855072 0.00000000 43.16564
## 26 6 82460167 rs115847999 C G 0.9855072 0.00000000 43.16564
## 27 6 82460379 rs116730626 G A 0.9855072 0.00000000 43.16564
## 28 6 82460650 rs149355051 G T 0.9855072 0.00000000 43.16564
## 29 6 82462126 rs76856243 T C 0.9855072 0.00000000 43.16564
## 30 6 82462603 rs6933239 C T 0.9855072 0.00000000 43.16564
## 31 6 82462992 rs6911216 A C 0.9855072 0.00000000 43.16564
## 32 6 82465617 rs6928806 A C 0.9855072 0.00000000 43.16564
## 33 6 82466574 rs74404091 C T 0.9855072 0.00000000 43.16564
## 34 6 82466715 rs79358384 G A 0.9855072 0.00000000 43.16564
## 35 6 82467200 rs77721025 A C 0.9855072 0.00000000 43.16564
## 36 6 82467706 rs116423281 A G 0.9855072 0.00000000 43.16564
## 37 6 82467953 rs142216660 T C 0.9855072 0.00000000 43.16564
## 38 6 82467988 rs140099919 A G 0.9855072 0.00000000 43.16564
## 39 6 82468668 rs78941585 G A 0.9855072 0.00000000 43.16564
## 40 6 82468774 rs75228268 A T 0.9855072 0.00000000 43.16564
## 41 6 82469116 rs77552654 T A 0.9855072 0.00000000 43.16564
## 42 6 82470649 rs149706908 G T 0.9855072 0.00000000 43.16564
## 43 6 82470651 rs144623191 T C 0.9855072 0.00000000 43.16564
## 44 6 82470859 rs192614396 C A 0.9855072 0.00000000 43.16564
## 45 6 82471119 rs151156468 C T 0.9855072 0.00000000 43.16564
## 46 6 82471338 rs140554533 A G 0.9855072 0.00000000 43.16564
## 47 6 82471381 rs150446760 C G 0.9855072 0.00000000 43.16564
## 48 6 82471679 rs138339145 T C 0.9855072 0.00000000 43.16564
## 49 6 82472140 rs142089466 T C 0.9855072 0.00000000 43.16564
## 50 6 82473676 rs147151928 G C 0.9855072 0.00000000 43.16564
## 51 6 82473679 rs140298049 C T 0.9855072 0.00000000 43.16564
## 52 6 82473688 rs149655171 A G 0.9855072 0.00000000 43.16564
## 53 6 82474468 rs146864360 T C 0.9855072 0.00000000 43.16564
## 54 6 82474648 rs140520501 C T 0.9855072 0.00000000 43.16564
## 55 6 82474867 rs150394443 T C 0.9855072 0.00000000 43.16564
## 56 6 82475320 rs143579868 A T 0.9855072 0.00000000 43.16564
## 57 6 82475696 rs115741750 C T 0.9855072 0.00000000 43.16564
## 58 6 82477810 rs77406497 T C 0.9855072 0.00000000 43.16564
## 59 6 82478378 rs115813741 T G 0.9855072 0.00000000 43.16564
## 60 6 82478650 rs78900243 G A 0.9855072 0.00000000 43.16564
## 61 6 82478870 rs76557363 T G 0.9855072 0.00000000 43.16564
## 62 6 82479264 rs77943130 T G 0.9855072 0.00000000 43.16564
## 63 6 82479546 rs115245280 A G 0.9855072 0.00000000 43.16564
## 64 6 82479664 rs75910615 A G 0.9855072 0.00000000 43.16564
## 65 6 82480370 rs76438934 A G 0.9855072 0.00000000 43.16564
## 66 6 82481986 rs115034961 A C 0.9855072 0.00000000 43.16564
## 67 6 82482183 rs79967481 G A 0.9855072 0.00000000 43.16564
## 68 6 82482254 rs116699765 T C 0.9855072 0.00000000 43.16564
## 69 6 82483125 rs78639250 T C 0.9855072 0.00000000 43.16564
## 70 6 82483522 rs75763492 A G 0.9855072 0.00000000 43.16564
## 71 6 82483898 rs76617768 T C 0.9855072 0.00000000 43.16564
## 72 6 82484287 rs76652063 C A 0.9855072 0.00000000 43.16564
## 73 6 82484758 rs114024754 A G 0.9855072 0.00000000 43.16564
## 74 6 82485370 rs79031996 C G 0.9855072 0.00000000 43.16564
## 75 6 82485916 rs77517398 C T 0.9855072 0.00000000 43.16564
## 76 6 82486863 rs80252551 A T 0.9855072 0.00000000 43.16564
## 77 6 82488997 rs114963095 G T 0.9855072 0.00000000 43.16564
## 78 6 82489053 rs77561386 C T 0.9855072 0.00000000 43.16564
## 79 6 82489164 rs76602861 A G 0.9855072 0.00000000 43.16564
## 80 6 82490178 rs79435750 A G 0.9855072 0.00000000 43.16564
## 81 6 82490367 rs80347071 C G 0.9855072 0.00000000 43.16564
## 82 6 82492170 rs74785531 C T 0.9855072 0.00000000 43.16564
## 83 6 82492233 rs74361967 C T 0.9855072 0.00000000 43.16564
## 84 6 82492321 rs75373608 A T 0.9855072 0.00000000 43.16564
## 85 10 64099069 rs74804993 G A 0.9826087 0.27726310 31.54649
## 86 16 13935176 rs1800067 A G 0.9550725 0.29435196 33.85516
## p
## 1 7.917990e-09
## 2 5.495605e-06
## 3 1.405911e-08
## 4 5.029621e-11
## 5 5.029621e-11
## 6 5.029621e-11
## 7 5.029621e-11
## 8 5.029621e-11
## 9 5.029621e-11
## 10 5.029621e-11
## 11 5.029621e-11
## 12 5.029621e-11
## 13 5.029621e-11
## 14 5.029621e-11
## 15 5.029621e-11
## 16 5.029621e-11
## 17 5.029621e-11
## 18 5.029621e-11
## 19 5.029621e-11
## 20 5.029621e-11
## 21 5.029621e-11
## 22 5.029621e-11
## 23 5.029621e-11
## 24 5.029621e-11
## 25 5.029621e-11
## 26 5.029621e-11
## 27 5.029621e-11
## 28 5.029621e-11
## 29 5.029621e-11
## 30 5.029621e-11
## 31 5.029621e-11
## 32 5.029621e-11
## 33 5.029621e-11
## 34 5.029621e-11
## 35 5.029621e-11
## 36 5.029621e-11
## 37 5.029621e-11
## 38 5.029621e-11
## 39 5.029621e-11
## 40 5.029621e-11
## 41 5.029621e-11
## 42 5.029621e-11
## 43 5.029621e-11
## 44 5.029621e-11
## 45 5.029621e-11
## 46 5.029621e-11
## 47 5.029621e-11
## 48 5.029621e-11
## 49 5.029621e-11
## 50 5.029621e-11
## 51 5.029621e-11
## 52 5.029621e-11
## 53 5.029621e-11
## 54 5.029621e-11
## 55 5.029621e-11
## 56 5.029621e-11
## 57 5.029621e-11
## 58 5.029621e-11
## 59 5.029621e-11
## 60 5.029621e-11
## 61 5.029621e-11
## 62 5.029621e-11
## 63 5.029621e-11
## 64 5.029621e-11
## 65 5.029621e-11
## 66 5.029621e-11
## 67 5.029621e-11
## 68 5.029621e-11
## 69 5.029621e-11
## 70 5.029621e-11
## 71 5.029621e-11
## 72 5.029621e-11
## 73 5.029621e-11
## 74 5.029621e-11
## 75 5.029621e-11
## 76 5.029621e-11
## 77 5.029621e-11
## 78 5.029621e-11
## 79 5.029621e-11
## 80 5.029621e-11
## 81 5.029621e-11
## 82 5.029621e-11
## 83 5.029621e-11
## 84 5.029621e-11
## 85 1.947216e-08
## 86 5.937145e-09manhattan(x = l_kvalH_filtered, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "L K-value H (LRT)", annotatePval = 1.84e-7)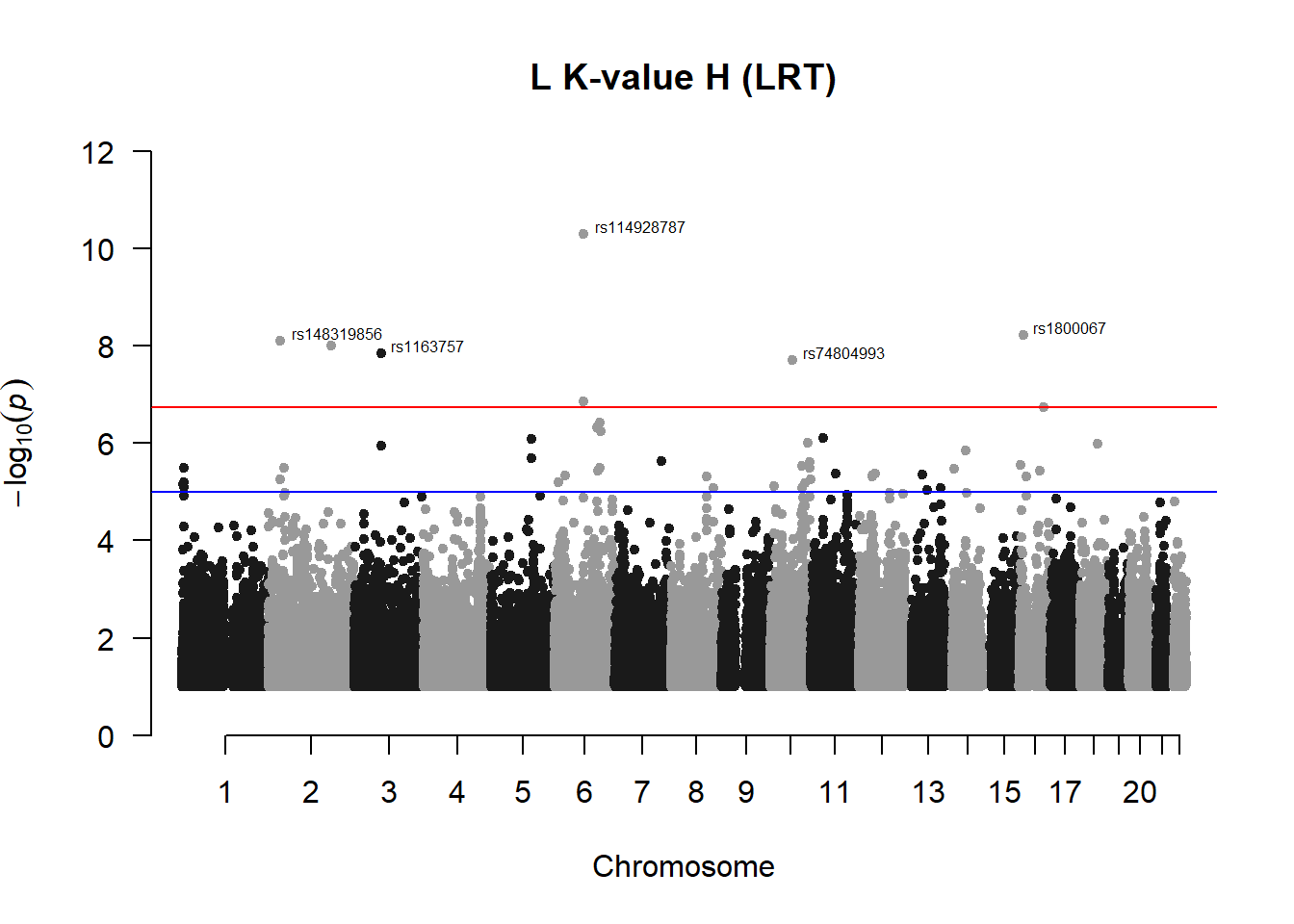
2. GWAS using lm and wald
l_KvalH_wald <- association.test(merged_nies_210818, nies_heritable_pheno$L.K.value.H, method="lm", test = "wald", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
l_KvalH_wald <- na.omit(l_KvalH_wald)
l_kvalH_Wfilt <- l_KvalH_wald %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
l_kvalH_Wmeff <- l_kvalH_Wfilt %>% filter(p < 1.84e-7)
l_kvalH_Wsig <- do.call(rbind, lapply(split(l_kvalH_Wmeff,l_kvalH_Wmeff$chr), function(x) {return(x[which.min(x$p),])}))
l_kvalH_Wres <- NULL
for (i in l_kvalH_sig$id) {
snpID <- i
snpCHR <- l_kvalH_Wfilt[l_kvalH_Wfilt$id == snpID,]$chr
snpPOS <- l_kvalH_Wfilt[l_kvalH_Wfilt$id ==snpID,]$pos
sig.peak <- l_kvalH_Wfilt %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
l_kvalH_Wres <- rbind(l_kvalH_Wres, sig.peak)
}
l_kvalH_Wres## chr pos id A1 A2 freqA2 beta sd
## 1 2 32321350 rs148319856 C A 0.9826087 2.792557 0.4665010
## 2 2 32367072 rs115270434 C A 0.9855072 2.480528 0.5178888
## 3 3 77739700 rs1163757 A G 0.9869565 3.221804 0.5333681
## 4 6 82425908 rs73481987 C T 0.9753623 1.917424 0.4054810
## 5 6 82442490 rs114928787 T C 0.9855072 3.173559 0.4545536
## 6 6 82442833 rs116172930 C A 0.9855072 3.173559 0.4545536
## 7 6 82443802 rs74436554 C T 0.9855072 3.173559 0.4545536
## 8 6 82444791 rs78056249 C G 0.9855072 3.173559 0.4545536
## 9 6 82445716 rs77941466 C T 0.9855072 3.173559 0.4545536
## 10 6 82447004 rs116461493 T C 0.9855072 3.173559 0.4545536
## 11 6 82447732 rs77666578 G A 0.9855072 3.173559 0.4545536
## 12 6 82451563 rs75189229 A G 0.9855072 3.173559 0.4545536
## 13 6 82453938 rs117178778 A G 0.9855072 3.173559 0.4545536
## 14 6 82454058 rs116246536 T G 0.9855072 3.173559 0.4545536
## 15 6 82454140 rs78427866 A G 0.9855072 3.173559 0.4545536
## 16 6 82454380 rs147257041 A G 0.9855072 3.173559 0.4545536
## 17 6 82454449 rs140711073 T C 0.9855072 3.173559 0.4545536
## 18 6 82454505 rs142248448 T C 0.9855072 3.173559 0.4545536
## 19 6 82454620 rs114442506 A G 0.9855072 3.173559 0.4545536
## 20 6 82456537 rs145558293 A G 0.9855072 3.173559 0.4545536
## 21 6 82458393 rs77391407 C T 0.9855072 3.173559 0.4545536
## 22 6 82458546 rs77103853 G C 0.9855072 3.173559 0.4545536
## 23 6 82459011 rs115096710 T C 0.9855072 3.173559 0.4545536
## 24 6 82459713 rs77245672 G C 0.9855072 3.173559 0.4545536
## 25 6 82459810 rs77599496 T A 0.9855072 3.173559 0.4545536
## 26 6 82459890 rs77598967 G A 0.9855072 3.173559 0.4545536
## 27 6 82460167 rs115847999 C G 0.9855072 3.173559 0.4545536
## 28 6 82460379 rs116730626 G A 0.9855072 3.173559 0.4545536
## 29 6 82460650 rs149355051 G T 0.9855072 3.173559 0.4545536
## 30 6 82462126 rs76856243 T C 0.9855072 3.173559 0.4545536
## 31 6 82462603 rs6933239 C T 0.9855072 3.173559 0.4545536
## 32 6 82462992 rs6911216 A C 0.9855072 3.173559 0.4545536
## 33 6 82465617 rs6928806 A C 0.9855072 3.173559 0.4545536
## 34 6 82466574 rs74404091 C T 0.9855072 3.173559 0.4545536
## 35 6 82466715 rs79358384 G A 0.9855072 3.173559 0.4545536
## 36 6 82467200 rs77721025 A C 0.9855072 3.173559 0.4545536
## 37 6 82467706 rs116423281 A G 0.9855072 3.173559 0.4545536
## 38 6 82467953 rs142216660 T C 0.9855072 3.173559 0.4545536
## 39 6 82467988 rs140099919 A G 0.9855072 3.173559 0.4545536
## 40 6 82468668 rs78941585 G A 0.9855072 3.173559 0.4545536
## 41 6 82468774 rs75228268 A T 0.9855072 3.173559 0.4545536
## 42 6 82469116 rs77552654 T A 0.9855072 3.173559 0.4545536
## 43 6 82470649 rs149706908 G T 0.9855072 3.173559 0.4545536
## 44 6 82470651 rs144623191 T C 0.9855072 3.173559 0.4545536
## 45 6 82470859 rs192614396 C A 0.9855072 3.173559 0.4545536
## 46 6 82471119 rs151156468 C T 0.9855072 3.173559 0.4545536
## 47 6 82471338 rs140554533 A G 0.9855072 3.173559 0.4545536
## 48 6 82471381 rs150446760 C G 0.9855072 3.173559 0.4545536
## 49 6 82471679 rs138339145 T C 0.9855072 3.173559 0.4545536
## 50 6 82472140 rs142089466 T C 0.9855072 3.173559 0.4545536
## 51 6 82473676 rs147151928 G C 0.9855072 3.173559 0.4545536
## 52 6 82473679 rs140298049 C T 0.9855072 3.173559 0.4545536
## 53 6 82473688 rs149655171 A G 0.9855072 3.173559 0.4545536
## 54 6 82474468 rs146864360 T C 0.9855072 3.173559 0.4545536
## 55 6 82474648 rs140520501 C T 0.9855072 3.173559 0.4545536
## 56 6 82474867 rs150394443 T C 0.9855072 3.173559 0.4545536
## 57 6 82475320 rs143579868 A T 0.9855072 3.173559 0.4545536
## 58 6 82475696 rs115741750 C T 0.9855072 3.173559 0.4545536
## 59 6 82477810 rs77406497 T C 0.9855072 3.173559 0.4545536
## 60 6 82478378 rs115813741 T G 0.9855072 3.173559 0.4545536
## 61 6 82478650 rs78900243 G A 0.9855072 3.173559 0.4545536
## 62 6 82478870 rs76557363 T G 0.9855072 3.173559 0.4545536
## 63 6 82479264 rs77943130 T G 0.9855072 3.173559 0.4545536
## 64 6 82479546 rs115245280 A G 0.9855072 3.173559 0.4545536
## 65 6 82479664 rs75910615 A G 0.9855072 3.173559 0.4545536
## 66 6 82480370 rs76438934 A G 0.9855072 3.173559 0.4545536
## 67 6 82481986 rs115034961 A C 0.9855072 3.173559 0.4545536
## 68 6 82482183 rs79967481 G A 0.9855072 3.173559 0.4545536
## 69 6 82482254 rs116699765 T C 0.9855072 3.173559 0.4545536
## 70 6 82483125 rs78639250 T C 0.9855072 3.173559 0.4545536
## 71 6 82483522 rs75763492 A G 0.9855072 3.173559 0.4545536
## 72 6 82483898 rs76617768 T C 0.9855072 3.173559 0.4545536
## 73 6 82484287 rs76652063 C A 0.9855072 3.173559 0.4545536
## 74 6 82484758 rs114024754 A G 0.9855072 3.173559 0.4545536
## 75 6 82485370 rs79031996 C G 0.9855072 3.173559 0.4545536
## 76 6 82485916 rs77517398 C T 0.9855072 3.173559 0.4545536
## 77 6 82486863 rs80252551 A T 0.9855072 3.173559 0.4545536
## 78 6 82488997 rs114963095 G T 0.9855072 3.173559 0.4545536
## 79 6 82489053 rs77561386 C T 0.9855072 3.173559 0.4545536
## 80 6 82489164 rs76602861 A G 0.9855072 3.173559 0.4545536
## 81 6 82490178 rs79435750 A G 0.9855072 3.173559 0.4545536
## 82 6 82490367 rs80347071 C G 0.9855072 3.173559 0.4545536
## 83 6 82492170 rs74785531 C T 0.9855072 3.173559 0.4545536
## 84 6 82492233 rs74361967 C T 0.9855072 3.173559 0.4545536
## 85 6 82492321 rs75373608 A T 0.9855072 3.173559 0.4545536
## 86 10 64099069 rs74804993 G A 0.9826087 2.637507 0.4654519
## 87 16 13935176 rs1800067 A G 0.9550725 1.635103 0.2819104
## p
## 1 5.442928e-09
## 2 2.495356e-06
## 3 4.020056e-09
## 4 3.311531e-06
## 5 1.530777e-11
## 6 1.530777e-11
## 7 1.530777e-11
## 8 1.530777e-11
## 9 1.530777e-11
## 10 1.530777e-11
## 11 1.530777e-11
## 12 1.530777e-11
## 13 1.530777e-11
## 14 1.530777e-11
## 15 1.530777e-11
## 16 1.530777e-11
## 17 1.530777e-11
## 18 1.530777e-11
## 19 1.530777e-11
## 20 1.530777e-11
## 21 1.530777e-11
## 22 1.530777e-11
## 23 1.530777e-11
## 24 1.530777e-11
## 25 1.530777e-11
## 26 1.530777e-11
## 27 1.530777e-11
## 28 1.530777e-11
## 29 1.530777e-11
## 30 1.530777e-11
## 31 1.530777e-11
## 32 1.530777e-11
## 33 1.530777e-11
## 34 1.530777e-11
## 35 1.530777e-11
## 36 1.530777e-11
## 37 1.530777e-11
## 38 1.530777e-11
## 39 1.530777e-11
## 40 1.530777e-11
## 41 1.530777e-11
## 42 1.530777e-11
## 43 1.530777e-11
## 44 1.530777e-11
## 45 1.530777e-11
## 46 1.530777e-11
## 47 1.530777e-11
## 48 1.530777e-11
## 49 1.530777e-11
## 50 1.530777e-11
## 51 1.530777e-11
## 52 1.530777e-11
## 53 1.530777e-11
## 54 1.530777e-11
## 55 1.530777e-11
## 56 1.530777e-11
## 57 1.530777e-11
## 58 1.530777e-11
## 59 1.530777e-11
## 60 1.530777e-11
## 61 1.530777e-11
## 62 1.530777e-11
## 63 1.530777e-11
## 64 1.530777e-11
## 65 1.530777e-11
## 66 1.530777e-11
## 67 1.530777e-11
## 68 1.530777e-11
## 69 1.530777e-11
## 70 1.530777e-11
## 71 1.530777e-11
## 72 1.530777e-11
## 73 1.530777e-11
## 74 1.530777e-11
## 75 1.530777e-11
## 76 1.530777e-11
## 77 1.530777e-11
## 78 1.530777e-11
## 79 1.530777e-11
## 80 1.530777e-11
## 81 1.530777e-11
## 82 1.530777e-11
## 83 1.530777e-11
## 84 1.530777e-11
## 85 1.530777e-11
## 86 3.102948e-08
## 87 1.512997e-08manhattan(x = l_kvalH_Wfilt, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "L K-value H (Wald)", annotatePval = 1.84e-7)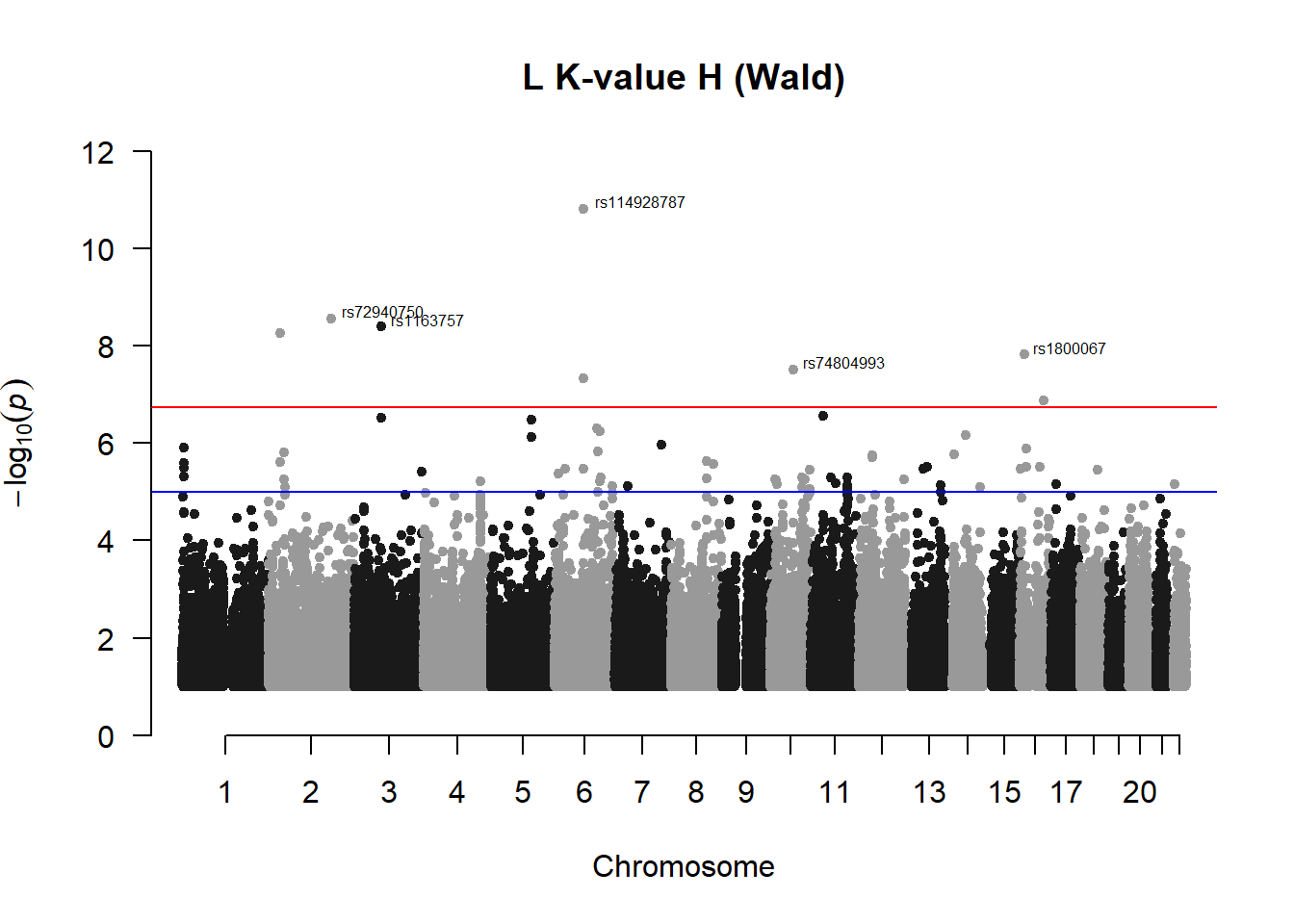
3. Test GWAS on R K-value V
r_kvalV_gwas <- association.test(merged_nies_210818, nies_heritable_pheno$R.K.value.V, method="lmm", test = "lrt", response = "quantitative", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
r_kvalV_gwas <- na.omit(r_kvalV_gwas)
r_kvalV_filtered <- r_kvalV_gwas %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
r_kvalV_meff <- r_kvalV_filtered %>% filter(p < 1.84e-7)
r_kvalV_sig <- do.call(rbind, lapply(split(r_kvalV_meff,r_kvalV_meff$chr), function(x) {return(x[which.min(x$p),])}))
r_kvalV_res <- NULL
for (i in r_kvalV_sig$id) {
snpID <- i
snpCHR <- r_kvalV_filtered[r_kvalV_filtered$id == snpID,]$chr
snpPOS <- r_kvalV_filtered[r_kvalV_filtered$id ==snpID,]$pos
sig.peak <- r_kvalV_filtered %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
r_kvalV_res <- rbind(r_kvalV_res, sig.peak)
}
r_kvalV_res## chr pos id A1 A2 freqA2 h2 LRT
## 1 1 205916566 rs72752923 C A 0.9782609 0.6943696 24.25721
## 2 1 205916729 rs16830364 G A 0.9782609 0.6943696 24.25721
## 3 1 205916931 rs60255052 C T 0.9811594 0.6398584 34.38041
## 4 1 205917041 rs60716530 C T 0.9782609 0.6943696 24.25721
## 5 1 205917115 rs60644810 G A 0.9782609 0.6943696 24.25721
## 6 1 205917248 rs35694044 A T 0.9782609 0.6943696 24.25721
## 7 1 205917652 rs16830370 T C 0.9782609 0.6943696 24.25721
## 8 1 205918853 rs16856462 C T 0.9782609 0.6943696 24.25721
## 9 1 205919195 rs16856468 G A 0.9782609 0.6943696 24.25721
## 10 1 205919303 rs60001177 T C 0.9782609 0.6943696 24.25721
## 11 1 205919401 rs12723666 A G 0.9782609 0.6943696 24.25721
## 12 1 205920201 rs16856470 T C 0.9811594 0.6398584 34.38041
## 13 1 205920404 rs16856473 A G 0.9811594 0.6398584 34.38041
## 14 1 205922403 rs68189466 G A 0.9797101 0.6457784 26.64533
## 15 1 205922720 rs35648260 G T 0.9797101 0.6457784 26.64533
## 16 1 205922775 rs35360452 G T 0.9797101 0.6457784 26.64533
## 17 1 205925474 rs12727528 A G 0.9797101 0.6457784 26.64533
## 18 1 205935183 rs1891309 G A 0.9782609 0.6459447 23.46813
## 19 1 205935330 rs1891310 T G 0.9782609 0.6459447 23.46813
## 20 1 205936240 rs1473537 T C 0.9739130 0.6448387 28.29076
## 21 1 205942026 rs72752928 T C 0.9782609 0.6459447 23.46813
## 22 1 205943691 rs66593238 T C 0.9782609 0.6459447 23.46813
## 23 1 205944627 rs9438407 T G 0.9782609 0.6459447 23.46813
## 24 1 205945388 rs12741299 T C 0.9782609 0.6459447 23.46813
## 25 1 205946647 rs34265780 T C 0.9782609 0.6459447 23.46813
## 26 4 119362510 rs112314510 A G 0.9855072 0.6858759 28.47085
## 27 4 119362552 rs111968273 T A 0.9855072 0.6858759 28.47085
## 28 4 119367677 rs113059419 A G 0.9869565 0.6769095 30.96024
## 29 4 119372303 rs112760591 G A 0.9855072 0.6858759 28.47085
## 30 4 119372480 rs111609258 C G 0.9855072 0.6858759 28.47085
## 31 4 119378224 rs147602981 G A 0.9855072 0.6858759 28.47085
## 32 4 119381167 rs143847948 A C 0.9855072 0.6858759 28.47085
## 33 4 119390257 rs113224532 T C 0.9855072 0.6858759 28.47085
## 34 4 119392279 rs117856633 C A 0.9855072 0.6858759 28.47085
## 35 4 119398681 rs113825627 C G 0.9855072 0.6858759 28.47085
## 36 4 119403429 rs112414671 A G 0.9869565 0.6769095 30.96024
## 37 4 119404439 rs183181597 A G 0.9855072 0.6858759 28.47085
## 38 4 119404927 rs113317707 C A 0.9855072 0.6858759 28.47085
## 39 4 119408454 rs184703141 T C 0.9855072 0.6858759 28.47085
## 40 15 57746156 rs143347058 A G 0.9898551 0.7970430 28.28088
## p
## 1 8.429104e-07
## 2 8.429104e-07
## 3 4.532633e-09
## 4 8.429104e-07
## 5 8.429104e-07
## 6 8.429104e-07
## 7 8.429104e-07
## 8 8.429104e-07
## 9 8.429104e-07
## 10 8.429104e-07
## 11 8.429104e-07
## 12 4.532633e-09
## 13 4.532633e-09
## 14 2.444408e-07
## 15 2.444408e-07
## 16 2.444408e-07
## 17 2.444408e-07
## 18 1.270004e-06
## 19 1.270004e-06
## 20 1.043931e-07
## 21 1.270004e-06
## 22 1.270004e-06
## 23 1.270004e-06
## 24 1.270004e-06
## 25 1.270004e-06
## 26 9.511997e-08
## 27 9.511997e-08
## 28 2.633682e-08
## 29 9.511997e-08
## 30 9.511997e-08
## 31 9.511997e-08
## 32 9.511997e-08
## 33 9.511997e-08
## 34 9.511997e-08
## 35 9.511997e-08
## 36 2.633682e-08
## 37 9.511997e-08
## 38 9.511997e-08
## 39 9.511997e-08
## 40 1.049275e-07manhattan(x = r_kvalV_filtered, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "R K-value V (LRT)", annotatePval = 1.84e-7)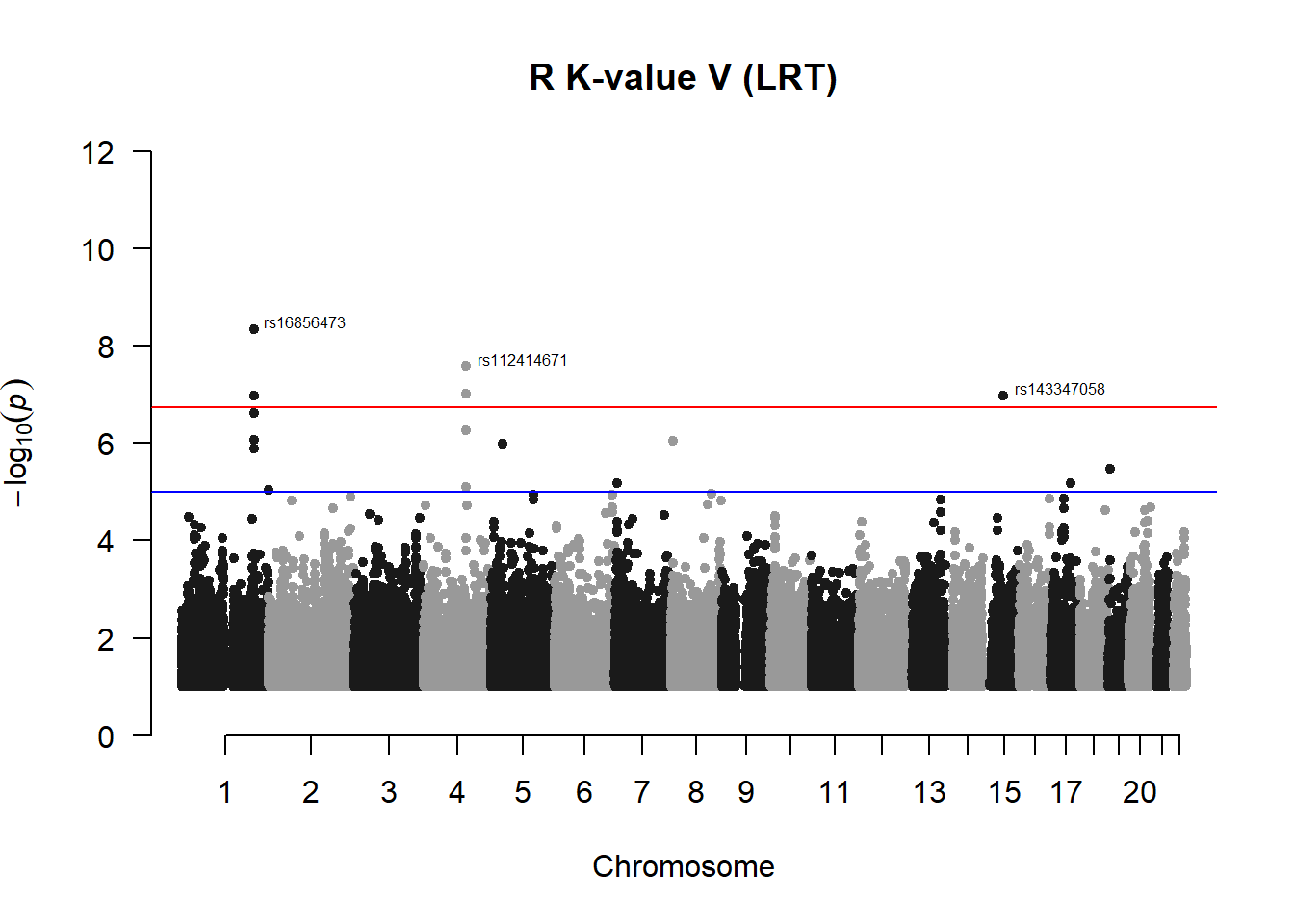
r_kvalV_wald <- association.test(merged_nies_210818, nies_heritable_pheno$R.K.value.V, method="lm", test = "wald", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
r_kvalV_wald <- na.omit(r_kvalV_wald)
r_kvalV_Wfilt <- r_kvalV_wald %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
r_kvalV_Wmeff <- r_kvalV_Wfilt %>% filter(p < 1.84e-7)
r_kvalV_Wsig <- do.call(rbind, lapply(split(r_kvalV_Wmeff,r_kvalV_Wmeff$chr), function(x) {return(x[which.min(x$p),])}))
r_kvalV_Wres <- NULL
for (i in r_kvalV_sig$id) {
snpID <- i
snpCHR <- r_kvalV_Wfilt[r_kvalV_Wfilt$id == snpID,]$chr
snpPOS <- r_kvalV_Wfilt[r_kvalV_Wfilt$id ==snpID,]$pos
sig.peak <- r_kvalV_Wfilt %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
r_kvalV_Wres <- rbind(r_kvalV_Wres, sig.peak)
}
r_kvalV_Wres## chr pos id A1 A2 freqA2 beta sd
## 1 1 205916566 rs72752923 C A 0.9782609 -2.094536 0.3862347
## 2 1 205916729 rs16830364 G A 0.9782609 -2.094536 0.3862347
## 3 1 205916931 rs60255052 C T 0.9811594 -2.694097 0.4018115
## 4 1 205917041 rs60716530 C T 0.9782609 -2.094536 0.3862347
## 5 1 205917115 rs60644810 G A 0.9782609 -2.094536 0.3862347
## 6 1 205917248 rs35694044 A T 0.9782609 -2.094536 0.3862347
## 7 1 205917652 rs16830370 T C 0.9782609 -2.094536 0.3862347
## 8 1 205918853 rs16856462 C T 0.9782609 -2.094536 0.3862347
## 9 1 205919195 rs16856468 G A 0.9782609 -2.094536 0.3862347
## 10 1 205919303 rs60001177 T C 0.9782609 -2.094536 0.3862347
## 11 1 205919401 rs12723666 A G 0.9782609 -2.094536 0.3862347
## 12 1 205920201 rs16856470 T C 0.9811594 -2.694097 0.4018115
## 13 1 205920404 rs16856473 A G 0.9811594 -2.694097 0.4018115
## 14 1 205922403 rs68189466 G A 0.9797101 -2.351940 0.3952791
## 15 1 205922720 rs35648260 G T 0.9797101 -2.351940 0.3952791
## 16 1 205922775 rs35360452 G T 0.9797101 -2.351940 0.3952791
## 17 1 205925474 rs12727528 A G 0.9797101 -2.351940 0.3952791
## 18 1 205935183 rs1891309 G A 0.9782609 -2.205056 0.3855569
## 19 1 205935330 rs1891310 T G 0.9782609 -2.205056 0.3855569
## 20 1 205936240 rs1473537 T C 0.9739130 -2.152699 0.3541437
## 21 1 205942026 rs72752928 T C 0.9782609 -2.205056 0.3855569
## 22 1 205943691 rs66593238 T C 0.9782609 -2.205056 0.3855569
## 23 1 205944627 rs9438407 T G 0.9782609 -2.205056 0.3855569
## 24 1 205945388 rs12741299 T C 0.9782609 -2.205056 0.3855569
## 25 1 205946647 rs34265780 T C 0.9782609 -2.205056 0.3855569
## 26 4 119362510 rs112314510 A G 0.9855072 -2.744140 0.4524155
## 27 4 119362552 rs111968273 T A 0.9855072 -2.744140 0.4524155
## 28 4 119367677 rs113059419 A G 0.9869565 -2.993797 0.4679999
## 29 4 119372303 rs112760591 G A 0.9855072 -2.744140 0.4524155
## 30 4 119372480 rs111609258 C G 0.9855072 -2.744140 0.4524155
## 31 4 119378224 rs147602981 G A 0.9855072 -2.744140 0.4524155
## 32 4 119381167 rs143847948 A C 0.9855072 -2.744140 0.4524155
## 33 4 119390257 rs113224532 T C 0.9855072 -2.744140 0.4524155
## 34 4 119392279 rs117856633 C A 0.9855072 -2.744140 0.4524155
## 35 4 119398681 rs113825627 C G 0.9855072 -2.744140 0.4524155
## 36 4 119403429 rs112414671 A G 0.9869565 -2.993797 0.4679999
## 37 4 119403485 rs185357281 A G 0.9869565 -2.297196 0.4814120
## 38 4 119404439 rs183181597 A G 0.9855072 -2.744140 0.4524155
## 39 4 119404927 rs113317707 C A 0.9855072 -2.744140 0.4524155
## 40 4 119408454 rs184703141 T C 0.9855072 -2.744140 0.4524155
## p
## 1 1.112376e-07
## 2 1.112376e-07
## 3 8.376034e-11
## 4 1.112376e-07
## 5 1.112376e-07
## 6 1.112376e-07
## 7 1.112376e-07
## 8 1.112376e-07
## 9 1.112376e-07
## 10 1.112376e-07
## 11 1.112376e-07
## 12 8.376034e-11
## 13 8.376034e-11
## 14 6.649744e-09
## 15 6.649744e-09
## 16 6.649744e-09
## 17 6.649744e-09
## 18 2.341944e-08
## 19 2.341944e-08
## 20 3.245978e-09
## 21 2.341944e-08
## 22 2.341944e-08
## 23 2.341944e-08
## 24 2.341944e-08
## 25 2.341944e-08
## 26 3.493475e-09
## 27 3.493475e-09
## 28 5.227359e-10
## 29 3.493475e-09
## 30 3.493475e-09
## 31 3.493475e-09
## 32 3.493475e-09
## 33 3.493475e-09
## 34 3.493475e-09
## 35 3.493475e-09
## 36 5.227359e-10
## 37 2.712563e-06
## 38 3.493475e-09
## 39 3.493475e-09
## 40 3.493475e-09manhattan(x = r_kvalV_Wfilt, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "R K-value V (Wald)", annotatePval = 1.84e-7)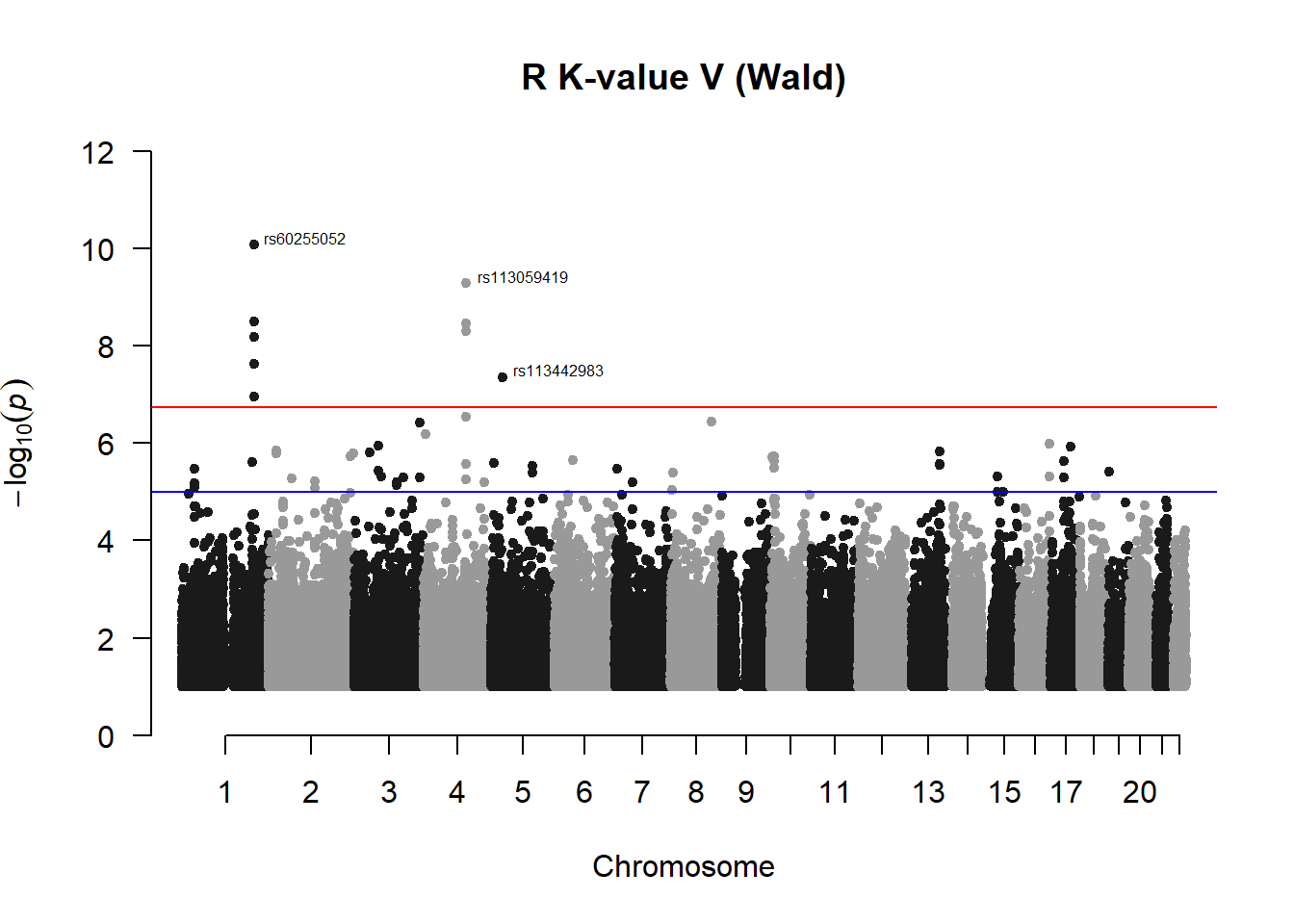
4. GWAS on component 3
pc3_gwas <- association.test(merged_nies_210818, nies_heritable_pheno$coord.Dim.3, method="lmm", test = "lrt", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
pc3_gwas <- na.omit(pc3_gwas)
pc3_filtered <- pc3_gwas %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
pc3_meff <- pc3_filtered %>% filter(p < 1.84e-7)
pc3_sig <- do.call(rbind, lapply(split(pc3_meff,pc3_meff$chr), function(x) {return(x[which.min(x$p),])}))
pc3_res <- NULL
for (i in pc3_sig$id) {
snpID <- i
snpCHR <- pc3_filtered[pc3_filtered$id == snpID,]$chr
snpPOS <- pc3_filtered[pc3_filtered$id ==snpID,]$pos
sig.peak <- pc3_filtered %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
pc3_res <- rbind(pc3_res, sig.peak)
}
pc3_res## chr pos id A1 A2 freqA2 h2 LRT
## 1 1 205916566 rs72752923 C A 0.9782609 0.10018393 32.70563
## 2 1 205916729 rs16830364 G A 0.9782609 0.10018393 32.70563
## 3 1 205916931 rs60255052 C T 0.9811594 0.06871623 39.43647
## 4 1 205917041 rs60716530 C T 0.9782609 0.10018393 32.70563
## 5 1 205917115 rs60644810 G A 0.9782609 0.10018393 32.70563
## 6 1 205917248 rs35694044 A T 0.9782609 0.10018393 32.70563
## 7 1 205917652 rs16830370 T C 0.9782609 0.10018393 32.70563
## 8 1 205918853 rs16856462 C T 0.9782609 0.10018393 32.70563
## 9 1 205919195 rs16856468 G A 0.9782609 0.10018393 32.70563
## 10 1 205919303 rs60001177 T C 0.9782609 0.10018393 32.70563
## 11 1 205919401 rs12723666 A G 0.9782609 0.10018393 32.70563
## 12 1 205920201 rs16856470 T C 0.9811594 0.06871623 39.43647
## 13 1 205920404 rs16856473 A G 0.9811594 0.06871623 39.43647
## 14 1 205922403 rs68189466 G A 0.9797101 0.07135981 40.62325
## 15 1 205922720 rs35648260 G T 0.9797101 0.07135981 40.62325
## 16 1 205922775 rs35360452 G T 0.9797101 0.07135981 40.62325
## 17 1 205925474 rs12727528 A G 0.9797101 0.07135981 40.62325
## 18 1 205935183 rs1891309 G A 0.9782609 0.06871632 34.26420
## 19 1 205935330 rs1891310 T G 0.9782609 0.06871632 34.26420
## 20 1 205936240 rs1473537 T C 0.9739130 0.10624855 27.50857
## 21 1 205942026 rs72752928 T C 0.9782609 0.06871632 34.26420
## 22 1 205943691 rs66593238 T C 0.9782609 0.06871632 34.26420
## 23 1 205944627 rs9438407 T G 0.9782609 0.06871632 34.26420
## 24 1 205945388 rs12741299 T C 0.9782609 0.06871632 34.26420
## 25 1 205946647 rs34265780 T C 0.9782609 0.06871632 34.26420
## 26 2 85686354 rs6759832 C T 0.9884058 0.14207905 36.56416
## 27 3 42006157 rs71315517 G C 0.9869565 0.06803235 36.98345
## 28 4 119362510 rs112314510 A G 0.9855072 0.07448922 41.35203
## 29 4 119362552 rs111968273 T A 0.9855072 0.07448922 41.35203
## 30 4 119367677 rs113059419 A G 0.9869565 0.04972603 45.15567
## 31 4 119372303 rs112760591 G A 0.9855072 0.07448922 41.35203
## 32 4 119372480 rs111609258 C G 0.9855072 0.07448922 41.35203
## 33 4 119378224 rs147602981 G A 0.9855072 0.07448922 41.35203
## 34 4 119381167 rs143847948 A C 0.9855072 0.07448922 41.35203
## 35 4 119390257 rs113224532 T C 0.9855072 0.07448922 41.35203
## 36 4 119392279 rs117856633 C A 0.9855072 0.07448922 41.35203
## 37 4 119398681 rs113825627 C G 0.9855072 0.07448922 41.35203
## 38 4 119403429 rs112414671 A G 0.9869565 0.04972603 45.15567
## 39 4 119403485 rs185357281 A G 0.9869565 0.06942631 28.75451
## 40 4 119404439 rs183181597 A G 0.9855072 0.07448922 41.35203
## 41 4 119404927 rs113317707 C A 0.9855072 0.07448922 41.35203
## 42 4 119408454 rs184703141 T C 0.9855072 0.07448922 41.35203
## 43 5 25293191 rs138955515 A G 0.9855072 0.07868229 31.90296
## 44 5 25300269 rs140158697 C T 0.9855072 0.07868229 31.90296
## 45 5 25331088 rs114133913 T A 0.9855072 0.07868229 31.90296
## 46 8 80306741 rs111864836 G T 0.9884058 0.10992188 30.94734
## 47 8 80306859 rs113627660 G C 0.9884058 0.10992188 30.94734
## 48 8 80307293 rs7845792 A G 0.9884058 0.10992188 30.94734
## 49 8 80307388 rs7845575 G A 0.9869565 0.12264506 24.21587
## 50 8 80307700 rs114654811 C T 0.9884058 0.10992188 30.94734
## 51 8 80307701 rs115278078 A T 0.9884058 0.10992188 30.94734
## 52 8 80307837 rs79018076 T C 0.9884058 0.10992188 30.94734
## 53 8 80308098 rs112501384 G C 0.9884058 0.10992188 30.94734
## 54 8 80309305 rs11996376 A G 0.9884058 0.10992188 30.94734
## 55 8 80309598 rs79431653 C G 0.9884058 0.10992188 30.94734
## 56 8 80309626 rs11996450 T C 0.9884058 0.10992188 30.94734
## 57 8 80310193 rs60741023 C G 0.9884058 0.10992188 30.94734
## 58 8 80310485 rs7844286 C T 0.9884058 0.10992188 30.94734
## 59 8 80310716 rs7844696 A T 0.9884058 0.10992188 30.94734
## 60 8 80310720 rs7826434 G A 0.9884058 0.10992188 30.94734
## 61 8 80311242 rs80031189 C T 0.9884058 0.10992188 30.94734
## 62 8 80311252 rs113823684 A G 0.9884058 0.10992188 30.94734
## 63 8 80311329 rs142907856 A G 0.9884058 0.10992188 30.94734
## 64 8 80311457 rs112582272 A G 0.9884058 0.10992188 30.94734
## 65 8 80311580 rs116773209 G A 0.9884058 0.10992188 30.94734
## 66 8 80311736 rs113314270 G T 0.9884058 0.10992188 30.94734
## 67 8 80311838 rs117693425 A G 0.9884058 0.10992188 30.94734
## 68 8 80312291 rs11988873 C T 0.9884058 0.10992188 30.94734
## 69 8 80312345 rs11988904 A T 0.9884058 0.10992188 30.94734
## 70 8 80312623 rs36051398 A C 0.9884058 0.10992188 30.94734
## 71 8 80313751 rs114426401 T C 0.9884058 0.10992188 30.94734
## 72 8 80314228 rs74981396 G A 0.9884058 0.10992188 30.94734
## 73 8 80317028 rs11998344 T G 0.9884058 0.10992188 30.94734
## 74 8 80317208 rs113244900 C T 0.9884058 0.10992188 30.94734
## 75 8 80318583 rs57246091 A G 0.9884058 0.10992188 30.94734
## 76 8 80318643 rs113901603 G A 0.9884058 0.10992188 30.94734
## 77 8 80319691 rs113718590 A G 0.9884058 0.10992188 30.94734
## 78 8 80320713 rs11987997 A G 0.9884058 0.10992188 30.94734
## 79 8 80321002 rs111527437 T A 0.9884058 0.10992188 30.94734
## 80 8 80321694 rs112815920 G C 0.9884058 0.10992188 30.94734
## 81 8 80323890 rs11998156 C A 0.9884058 0.10992188 30.94734
## 82 8 80323964 rs16907573 G A 0.9884058 0.10992188 30.94734
## 83 8 80324156 rs111257240 C T 0.9884058 0.10992188 30.94734
## 84 8 80325047 rs113437388 T C 0.9884058 0.10992188 30.94734
## 85 8 80326033 rs111370206 G A 0.9884058 0.10992188 30.94734
## 86 8 80326935 rs147955676 A G 0.9884058 0.10992188 30.94734
## 87 8 80327442 rs7841095 C T 0.9884058 0.10992188 30.94734
## 88 8 80327549 rs7822828 C A 0.9884058 0.10992188 30.94734
## 89 8 80327732 rs7823481 C G 0.9884058 0.10992188 30.94734
## 90 8 80328450 rs11990649 A G 0.9884058 0.10992188 30.94734
## 91 8 80329801 rs56960849 A G 0.9884058 0.10992188 30.94734
## 92 8 80330197 rs11995624 C T 0.9884058 0.10992188 30.94734
## 93 8 80330467 rs7837761 A G 0.9869565 0.12264506 24.21587
## 94 8 80332180 rs113042618 T C 0.9884058 0.10992188 30.94734
## 95 8 80332423 rs7001142 G T 0.9884058 0.10992188 30.94734
## 96 8 80332520 rs6980915 G A 0.9884058 0.10992188 30.94734
## 97 8 80332901 rs6473220 T C 0.9884058 0.10992188 30.94734
## 98 8 80332907 rs6473221 A G 0.9884058 0.10992188 30.94734
## 99 8 80333316 rs7835862 C T 0.9884058 0.10992188 30.94734
## 100 8 80333340 rs7817696 A G 0.9884058 0.10992188 30.94734
## 101 8 80333506 rs7817634 T C 0.9884058 0.10992188 30.94734
## 102 8 80333599 rs57419267 C T 0.9884058 0.10992188 30.94734
## 103 8 80333823 rs59130219 C T 0.9884058 0.10992188 30.94734
## 104 8 80334098 rs73691385 T A 0.9884058 0.10992188 30.94734
## 105 8 80334328 rs7840559 C T 0.9884058 0.10992188 30.94734
## 106 8 80334334 rs7822177 G A 0.9884058 0.10992188 30.94734
## 107 8 80334582 rs111631456 A G 0.9884058 0.10992188 30.94734
## 108 8 80334982 rs58109903 T C 0.9884058 0.10992188 30.94734
## 109 8 80335583 rs73691388 G A 0.9884058 0.10992188 30.94734
## 110 8 80336124 rs7837108 T C 0.9884058 0.10992188 30.94734
## 111 8 80336300 rs7840857 T G 0.9884058 0.10992188 30.94734
## 112 8 80336310 rs7840861 T G 0.9884058 0.10992188 30.94734
## 113 8 80337292 rs111888115 T C 0.9884058 0.10992188 30.94734
## 114 8 80337308 rs56871558 T C 0.9884058 0.10992188 30.94734
## 115 8 80337408 rs56116331 A G 0.9884058 0.10992188 30.94734
## 116 8 80337443 rs58592997 T C 0.9884058 0.10992188 30.94734
## 117 8 80337629 rs76783901 A G 0.9884058 0.10992188 30.94734
## 118 8 80337634 rs74468856 G A 0.9884058 0.10992188 30.94734
## 119 8 80337985 rs73691394 C A 0.9884058 0.10992188 30.94734
## 120 8 80338076 rs73691395 T C 0.9884058 0.10992188 30.94734
## 121 8 80338219 rs73691397 C G 0.9884058 0.10992188 30.94734
## 122 8 80338255 rs73691398 G A 0.9884058 0.10992188 30.94734
## 123 8 80338835 rs58958046 A G 0.9884058 0.10992188 30.94734
## 124 8 80338910 rs58786240 C T 0.9884058 0.10992188 30.94734
## 125 8 80338955 rs76867788 C T 0.9884058 0.10992188 30.94734
## 126 8 80339017 rs73691399 T C 0.9884058 0.10992188 30.94734
## 127 8 80339217 rs73691400 C G 0.9884058 0.10992188 30.94734
## 128 8 80339959 rs61233095 C A 0.9884058 0.10992188 30.94734
## 129 8 80340238 rs56408864 A T 0.9884058 0.10992188 30.94734
## 130 8 80340749 rs7826667 A G 0.9884058 0.10992188 30.94734
## 131 8 80341236 rs7000592 G C 0.9884058 0.10992188 30.94734
## 132 12 5987972 rs142100550 T G 0.9884058 0.06670431 28.13313
## 133 14 77833818 rs146124823 G A 0.9898551 0.14932850 29.35113
## p
## 1 1.072260e-08
## 2 1.072260e-08
## 3 3.389072e-10
## 4 1.072260e-08
## 5 1.072260e-08
## 6 1.072260e-08
## 7 1.072260e-08
## 8 1.072260e-08
## 9 1.072260e-08
## 10 1.072260e-08
## 11 1.072260e-08
## 12 3.389072e-10
## 13 3.389072e-10
## 14 1.845969e-10
## 15 1.845969e-10
## 16 1.845969e-10
## 17 1.845969e-10
## 18 4.811508e-09
## 19 4.811508e-09
## 20 1.563996e-07
## 21 4.811508e-09
## 22 4.811508e-09
## 23 4.811508e-09
## 24 4.811508e-09
## 25 4.811508e-09
## 26 1.477230e-09
## 27 1.191362e-09
## 28 1.271391e-10
## 29 1.271391e-10
## 30 1.819783e-11
## 31 1.271391e-10
## 32 1.271391e-10
## 33 1.271391e-10
## 34 1.271391e-10
## 35 1.271391e-10
## 36 1.271391e-10
## 37 1.271391e-10
## 38 1.819783e-11
## 39 8.215837e-08
## 40 1.271391e-10
## 41 1.271391e-10
## 42 1.271391e-10
## 43 1.620700e-08
## 44 1.620700e-08
## 45 1.620700e-08
## 46 2.651253e-08
## 47 2.651253e-08
## 48 2.651253e-08
## 49 8.611958e-07
## 50 2.651253e-08
## 51 2.651253e-08
## 52 2.651253e-08
## 53 2.651253e-08
## 54 2.651253e-08
## 55 2.651253e-08
## 56 2.651253e-08
## 57 2.651253e-08
## 58 2.651253e-08
## 59 2.651253e-08
## 60 2.651253e-08
## 61 2.651253e-08
## 62 2.651253e-08
## 63 2.651253e-08
## 64 2.651253e-08
## 65 2.651253e-08
## 66 2.651253e-08
## 67 2.651253e-08
## 68 2.651253e-08
## 69 2.651253e-08
## 70 2.651253e-08
## 71 2.651253e-08
## 72 2.651253e-08
## 73 2.651253e-08
## 74 2.651253e-08
## 75 2.651253e-08
## 76 2.651253e-08
## 77 2.651253e-08
## 78 2.651253e-08
## 79 2.651253e-08
## 80 2.651253e-08
## 81 2.651253e-08
## 82 2.651253e-08
## 83 2.651253e-08
## 84 2.651253e-08
## 85 2.651253e-08
## 86 2.651253e-08
## 87 2.651253e-08
## 88 2.651253e-08
## 89 2.651253e-08
## 90 2.651253e-08
## 91 2.651253e-08
## 92 2.651253e-08
## 93 8.611958e-07
## 94 2.651253e-08
## 95 2.651253e-08
## 96 2.651253e-08
## 97 2.651253e-08
## 98 2.651253e-08
## 99 2.651253e-08
## 100 2.651253e-08
## 101 2.651253e-08
## 102 2.651253e-08
## 103 2.651253e-08
## 104 2.651253e-08
## 105 2.651253e-08
## 106 2.651253e-08
## 107 2.651253e-08
## 108 2.651253e-08
## 109 2.651253e-08
## 110 2.651253e-08
## 111 2.651253e-08
## 112 2.651253e-08
## 113 2.651253e-08
## 114 2.651253e-08
## 115 2.651253e-08
## 116 2.651253e-08
## 117 2.651253e-08
## 118 2.651253e-08
## 119 2.651253e-08
## 120 2.651253e-08
## 121 2.651253e-08
## 122 2.651253e-08
## 123 2.651253e-08
## 124 2.651253e-08
## 125 2.651253e-08
## 126 2.651253e-08
## 127 2.651253e-08
## 128 2.651253e-08
## 129 2.651253e-08
## 130 2.651253e-08
## 131 2.651253e-08
## 132 1.132505e-07
## 133 6.038130e-08manhattan(x = pc3_filtered, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "Component 3 (LRT)", annotatePval = 1.84e-7)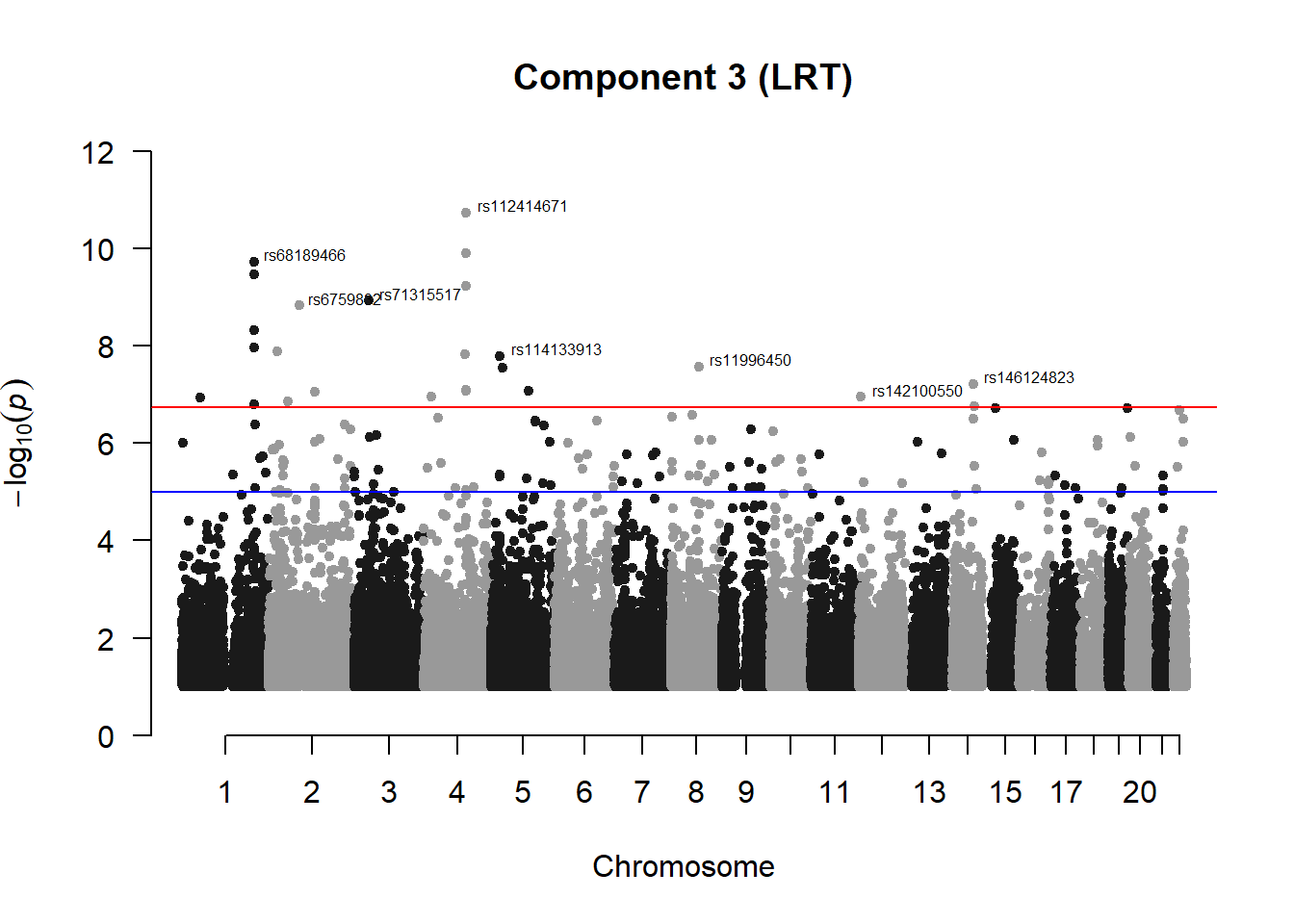
pc3_wald <- association.test(merged_nies_210818, nies_heritable_pheno$coord.Dim.3, method="lm", test = "wald", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
pc3_wald <- na.omit(pc3_wald)
pc3_Wfilt <- pc3_wald %>% filter(-log10(p)>1) %>% filter(freqA2 < 0.99)
pc3_Wmeff <- pc3_Wfilt %>% filter(p < 1.84e-7)
pc3_Wsig <- do.call(rbind, lapply(split(pc3_Wmeff,pc3_Wmeff$chr), function(x) {return(x[which.min(x$p),])}))
pc3_Wres <- NULL
for (i in pc3_sig$id) {
snpID <- i
snpCHR <- pc3_Wfilt[pc3_Wfilt$id == snpID,]$chr
snpPOS <- pc3_Wfilt[pc3_Wfilt$id ==snpID,]$pos
sig.peak <- pc3_Wfilt %>%
filter(chr == snpCHR) %>%
filter(pos > snpPOS - 50000) %>%
filter(pos < snpPOS + 50000) %>%
filter(p < 1e-5)
pc3_Wres <- rbind(pc3_Wres, sig.peak)
}
pc3_Wres## chr pos id A1 A2 freqA2 beta sd
## 1 1 205916566 rs72752923 C A 0.9782609 -2.667653 0.4393435
## 2 1 205916729 rs16830364 G A 0.9782609 -2.667653 0.4393435
## 3 1 205916931 rs60255052 C T 0.9811594 -3.088674 0.4617708
## 4 1 205917041 rs60716530 C T 0.9782609 -2.667653 0.4393435
## 5 1 205917115 rs60644810 G A 0.9782609 -2.667653 0.4393435
## 6 1 205917248 rs35694044 A T 0.9782609 -2.667653 0.4393435
## 7 1 205917652 rs16830370 T C 0.9782609 -2.667653 0.4393435
## 8 1 205918853 rs16856462 C T 0.9782609 -2.667653 0.4393435
## 9 1 205919195 rs16856468 G A 0.9782609 -2.667653 0.4393435
## 10 1 205919303 rs60001177 T C 0.9782609 -2.667653 0.4393435
## 11 1 205919401 rs12723666 A G 0.9782609 -2.667653 0.4393435
## 12 1 205920201 rs16856470 T C 0.9811594 -3.088674 0.4617708
## 13 1 205920404 rs16856473 A G 0.9811594 -3.088674 0.4617708
## 14 1 205922403 rs68189466 G A 0.9797101 -3.038821 0.4478525
## 15 1 205922720 rs35648260 G T 0.9797101 -3.038821 0.4478525
## 16 1 205922775 rs35360452 G T 0.9797101 -3.038821 0.4478525
## 17 1 205925474 rs12727528 A G 0.9797101 -3.038821 0.4478525
## 18 1 205935183 rs1891309 G A 0.9782609 -2.744145 0.4392695
## 19 1 205935330 rs1891310 T G 0.9782609 -2.744145 0.4392695
## 20 1 205936240 rs1473537 T C 0.9739130 -2.288989 0.4100258
## 21 1 205942026 rs72752928 T C 0.9782609 -2.744145 0.4392695
## 22 1 205943691 rs66593238 T C 0.9782609 -2.744145 0.4392695
## 23 1 205944627 rs9438407 T G 0.9782609 -2.744145 0.4392695
## 24 1 205945388 rs12741299 T C 0.9782609 -2.744145 0.4392695
## 25 1 205946647 rs34265780 T C 0.9782609 -2.744145 0.4392695
## 26 2 85686354 rs6759832 C T 0.9884058 -4.239789 0.6711844
## 27 3 42006157 rs71315517 G C 0.9869565 -3.493108 0.5392500
## 28 4 119362510 rs112314510 A G 0.9855072 -3.508836 0.5130453
## 29 4 119362552 rs111968273 T A 0.9855072 -3.508836 0.5130453
## 30 4 119367677 rs113059419 A G 0.9869565 -3.801920 0.5304818
## 31 4 119372303 rs112760591 G A 0.9855072 -3.508836 0.5130453
## 32 4 119372480 rs111609258 C G 0.9855072 -3.508836 0.5130453
## 33 4 119378224 rs147602981 G A 0.9855072 -3.508836 0.5130453
## 34 4 119381167 rs143847948 A C 0.9855072 -3.508836 0.5130453
## 35 4 119390257 rs113224532 T C 0.9855072 -3.508836 0.5130453
## 36 4 119392279 rs117856633 C A 0.9855072 -3.508836 0.5130453
## 37 4 119398681 rs113825627 C G 0.9855072 -3.508836 0.5130453
## 38 4 119403429 rs112414671 A G 0.9869565 -3.801920 0.5304818
## 39 4 119403485 rs185357281 A G 0.9869565 -3.132726 0.5454914
## 40 4 119404439 rs183181597 A G 0.9855072 -3.508836 0.5130453
## 41 4 119404927 rs113317707 C A 0.9855072 -3.508836 0.5130453
## 42 4 119408454 rs184703141 T C 0.9855072 -3.508836 0.5130453
## 43 5 25293191 rs138955515 A G 0.9855072 -3.122567 0.5186110
## 44 5 25300269 rs140158697 C T 0.9855072 -3.122567 0.5186110
## 45 5 25331088 rs114133913 T A 0.9855072 -3.122567 0.5186110
## 46 8 80306741 rs111864836 G T 0.9884058 -3.735501 0.6324799
## 47 8 80306859 rs113627660 G C 0.9884058 -3.735501 0.6324799
## 48 8 80307293 rs7845792 A G 0.9884058 -3.735501 0.6324799
## 49 8 80307388 rs7845575 G A 0.9869565 -3.159137 0.6030281
## 50 8 80307700 rs114654811 C T 0.9884058 -3.735501 0.6324799
## 51 8 80307701 rs115278078 A T 0.9884058 -3.735501 0.6324799
## 52 8 80307837 rs79018076 T C 0.9884058 -3.735501 0.6324799
## 53 8 80308098 rs112501384 G C 0.9884058 -3.735501 0.6324799
## 54 8 80309305 rs11996376 A G 0.9884058 -3.735501 0.6324799
## 55 8 80309598 rs79431653 C G 0.9884058 -3.735501 0.6324799
## 56 8 80309626 rs11996450 T C 0.9884058 -3.735501 0.6324799
## 57 8 80310193 rs60741023 C G 0.9884058 -3.735501 0.6324799
## 58 8 80310485 rs7844286 C T 0.9884058 -3.735501 0.6324799
## 59 8 80310716 rs7844696 A T 0.9884058 -3.735501 0.6324799
## 60 8 80310720 rs7826434 G A 0.9884058 -3.735501 0.6324799
## 61 8 80311242 rs80031189 C T 0.9884058 -3.735501 0.6324799
## 62 8 80311252 rs113823684 A G 0.9884058 -3.735501 0.6324799
## 63 8 80311329 rs142907856 A G 0.9884058 -3.735501 0.6324799
## 64 8 80311457 rs112582272 A G 0.9884058 -3.735501 0.6324799
## 65 8 80311580 rs116773209 G A 0.9884058 -3.735501 0.6324799
## 66 8 80311736 rs113314270 G T 0.9884058 -3.735501 0.6324799
## 67 8 80311838 rs117693425 A G 0.9884058 -3.735501 0.6324799
## 68 8 80312291 rs11988873 C T 0.9884058 -3.735501 0.6324799
## 69 8 80312345 rs11988904 A T 0.9884058 -3.735501 0.6324799
## 70 8 80312623 rs36051398 A C 0.9884058 -3.735501 0.6324799
## 71 8 80313751 rs114426401 T C 0.9884058 -3.735501 0.6324799
## 72 8 80314228 rs74981396 G A 0.9884058 -3.735501 0.6324799
## 73 8 80317028 rs11998344 T G 0.9884058 -3.735501 0.6324799
## 74 8 80317208 rs113244900 C T 0.9884058 -3.735501 0.6324799
## 75 8 80318583 rs57246091 A G 0.9884058 -3.735501 0.6324799
## 76 8 80318643 rs113901603 G A 0.9884058 -3.735501 0.6324799
## 77 8 80319691 rs113718590 A G 0.9884058 -3.735501 0.6324799
## 78 8 80320713 rs11987997 A G 0.9884058 -3.735501 0.6324799
## 79 8 80321002 rs111527437 T A 0.9884058 -3.735501 0.6324799
## 80 8 80321694 rs112815920 G C 0.9884058 -3.735501 0.6324799
## 81 8 80323890 rs11998156 C A 0.9884058 -3.735501 0.6324799
## 82 8 80323964 rs16907573 G A 0.9884058 -3.735501 0.6324799
## 83 8 80324156 rs111257240 C T 0.9884058 -3.735501 0.6324799
## 84 8 80325047 rs113437388 T C 0.9884058 -3.735501 0.6324799
## 85 8 80326033 rs111370206 G A 0.9884058 -3.735501 0.6324799
## 86 8 80326935 rs147955676 A G 0.9884058 -3.735501 0.6324799
## 87 8 80327442 rs7841095 C T 0.9884058 -3.735501 0.6324799
## 88 8 80327549 rs7822828 C A 0.9884058 -3.735501 0.6324799
## 89 8 80327732 rs7823481 C G 0.9884058 -3.735501 0.6324799
## 90 8 80328450 rs11990649 A G 0.9884058 -3.735501 0.6324799
## 91 8 80329801 rs56960849 A G 0.9884058 -3.735501 0.6324799
## 92 8 80330197 rs11995624 C T 0.9884058 -3.735501 0.6324799
## 93 8 80330467 rs7837761 A G 0.9869565 -3.159137 0.6030281
## 94 8 80332180 rs113042618 T C 0.9884058 -3.735501 0.6324799
## 95 8 80332423 rs7001142 G T 0.9884058 -3.735501 0.6324799
## 96 8 80332520 rs6980915 G A 0.9884058 -3.735501 0.6324799
## 97 8 80332901 rs6473220 T C 0.9884058 -3.735501 0.6324799
## 98 8 80332907 rs6473221 A G 0.9884058 -3.735501 0.6324799
## 99 8 80333316 rs7835862 C T 0.9884058 -3.735501 0.6324799
## 100 8 80333340 rs7817696 A G 0.9884058 -3.735501 0.6324799
## 101 8 80333506 rs7817634 T C 0.9884058 -3.735501 0.6324799
## 102 8 80333599 rs57419267 C T 0.9884058 -3.735501 0.6324799
## 103 8 80333823 rs59130219 C T 0.9884058 -3.735501 0.6324799
## 104 8 80334098 rs73691385 T A 0.9884058 -3.735501 0.6324799
## 105 8 80334328 rs7840559 C T 0.9884058 -3.735501 0.6324799
## 106 8 80334334 rs7822177 G A 0.9884058 -3.735501 0.6324799
## 107 8 80334582 rs111631456 A G 0.9884058 -3.735501 0.6324799
## 108 8 80334982 rs58109903 T C 0.9884058 -3.735501 0.6324799
## 109 8 80335583 rs73691388 G A 0.9884058 -3.735501 0.6324799
## 110 8 80336124 rs7837108 T C 0.9884058 -3.735501 0.6324799
## 111 8 80336300 rs7840857 T G 0.9884058 -3.735501 0.6324799
## 112 8 80336310 rs7840861 T G 0.9884058 -3.735501 0.6324799
## 113 8 80337292 rs111888115 T C 0.9884058 -3.735501 0.6324799
## 114 8 80337308 rs56871558 T C 0.9884058 -3.735501 0.6324799
## 115 8 80337408 rs56116331 A G 0.9884058 -3.735501 0.6324799
## 116 8 80337443 rs58592997 T C 0.9884058 -3.735501 0.6324799
## 117 8 80337629 rs76783901 A G 0.9884058 -3.735501 0.6324799
## 118 8 80337634 rs74468856 G A 0.9884058 -3.735501 0.6324799
## 119 8 80337985 rs73691394 C A 0.9884058 -3.735501 0.6324799
## 120 8 80338076 rs73691395 T C 0.9884058 -3.735501 0.6324799
## 121 8 80338219 rs73691397 C G 0.9884058 -3.735501 0.6324799
## 122 8 80338255 rs73691398 G A 0.9884058 -3.735501 0.6324799
## 123 8 80338835 rs58958046 A G 0.9884058 -3.735501 0.6324799
## 124 8 80338910 rs58786240 C T 0.9884058 -3.735501 0.6324799
## 125 8 80338955 rs76867788 C T 0.9884058 -3.735501 0.6324799
## 126 8 80339017 rs73691399 T C 0.9884058 -3.735501 0.6324799
## 127 8 80339217 rs73691400 C G 0.9884058 -3.735501 0.6324799
## 128 8 80339959 rs61233095 C A 0.9884058 -3.735501 0.6324799
## 129 8 80340238 rs56408864 A T 0.9884058 -3.735501 0.6324799
## 130 8 80340749 rs7826667 A G 0.9884058 -3.735501 0.6324799
## 131 8 80341236 rs7000592 G C 0.9884058 -3.735501 0.6324799
## 132 12 5987972 rs142100550 T G 0.9884058 -3.606900 0.6340382
## 133 14 77833818 rs146124823 G A 0.9898551 -3.360562 0.5957111
## p
## 1 3.370520e-09
## 2 3.370520e-09
## 3 9.233125e-11
## 4 3.370520e-09
## 5 3.370520e-09
## 6 3.370520e-09
## 7 3.370520e-09
## 8 3.370520e-09
## 9 3.370520e-09
## 10 3.370520e-09
## 11 3.370520e-09
## 12 9.233125e-11
## 13 9.233125e-11
## 14 5.137812e-11
## 15 5.137812e-11
## 16 5.137812e-11
## 17 5.137812e-11
## 18 1.246023e-09
## 19 1.246023e-09
## 20 4.843124e-08
## 21 1.246023e-09
## 22 1.246023e-09
## 23 1.246023e-09
## 24 1.246023e-09
## 25 1.246023e-09
## 26 8.331221e-10
## 27 3.254345e-10
## 28 3.693930e-11
## 29 3.693930e-11
## 30 4.777543e-12
## 31 3.693930e-11
## 32 3.693930e-11
## 33 3.693930e-11
## 34 3.693930e-11
## 35 3.693930e-11
## 36 3.693930e-11
## 37 3.693930e-11
## 38 4.777543e-12
## 39 2.060641e-08
## 40 3.693930e-11
## 41 3.693930e-11
## 42 3.693930e-11
## 43 4.482416e-09
## 44 4.482416e-09
## 45 4.482416e-09
## 46 8.475295e-09
## 47 8.475295e-09
## 48 8.475295e-09
## 49 2.836489e-07
## 50 8.475295e-09
## 51 8.475295e-09
## 52 8.475295e-09
## 53 8.475295e-09
## 54 8.475295e-09
## 55 8.475295e-09
## 56 8.475295e-09
## 57 8.475295e-09
## 58 8.475295e-09
## 59 8.475295e-09
## 60 8.475295e-09
## 61 8.475295e-09
## 62 8.475295e-09
## 63 8.475295e-09
## 64 8.475295e-09
## 65 8.475295e-09
## 66 8.475295e-09
## 67 8.475295e-09
## 68 8.475295e-09
## 69 8.475295e-09
## 70 8.475295e-09
## 71 8.475295e-09
## 72 8.475295e-09
## 73 8.475295e-09
## 74 8.475295e-09
## 75 8.475295e-09
## 76 8.475295e-09
## 77 8.475295e-09
## 78 8.475295e-09
## 79 8.475295e-09
## 80 8.475295e-09
## 81 8.475295e-09
## 82 8.475295e-09
## 83 8.475295e-09
## 84 8.475295e-09
## 85 8.475295e-09
## 86 8.475295e-09
## 87 8.475295e-09
## 88 8.475295e-09
## 89 8.475295e-09
## 90 8.475295e-09
## 91 8.475295e-09
## 92 8.475295e-09
## 93 2.836489e-07
## 94 8.475295e-09
## 95 8.475295e-09
## 96 8.475295e-09
## 97 8.475295e-09
## 98 8.475295e-09
## 99 8.475295e-09
## 100 8.475295e-09
## 101 8.475295e-09
## 102 8.475295e-09
## 103 8.475295e-09
## 104 8.475295e-09
## 105 8.475295e-09
## 106 8.475295e-09
## 107 8.475295e-09
## 108 8.475295e-09
## 109 8.475295e-09
## 110 8.475295e-09
## 111 8.475295e-09
## 112 8.475295e-09
## 113 8.475295e-09
## 114 8.475295e-09
## 115 8.475295e-09
## 116 8.475295e-09
## 117 8.475295e-09
## 118 8.475295e-09
## 119 8.475295e-09
## 120 8.475295e-09
## 121 8.475295e-09
## 122 8.475295e-09
## 123 8.475295e-09
## 124 8.475295e-09
## 125 8.475295e-09
## 126 8.475295e-09
## 127 8.475295e-09
## 128 8.475295e-09
## 129 8.475295e-09
## 130 8.475295e-09
## 131 8.475295e-09
## 132 2.755790e-08
## 133 3.549908e-08manhattan(x = pc3_Wfilt, chr = "chr", bp = "pos", p = "p", snp = "id", ylim = c(0,12), genomewideline = -log10(1.84e-7), main = "Component 3 (Wald)", annotatePval = 1.84e-7)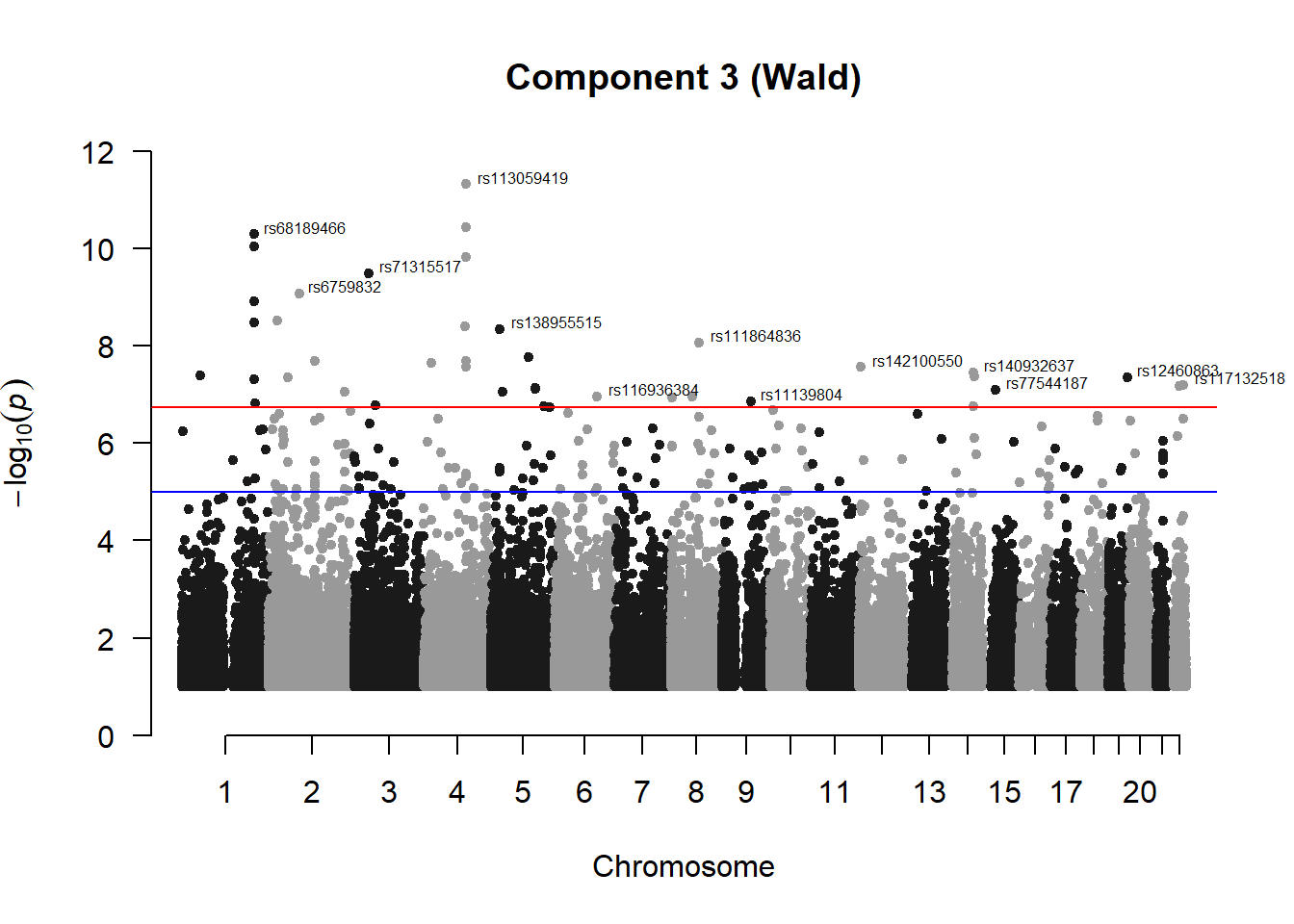
Wald test appears to identify more significant SNPs
5. Generate example Q-Q plots.
For componenet 3
pc3_rare_out <- pc3_gwas %>% filter(freqA2 < 0.99)
qqplot.pvalues(pc3_rare_out$p, main = "Component 3 QQ plot of p-values")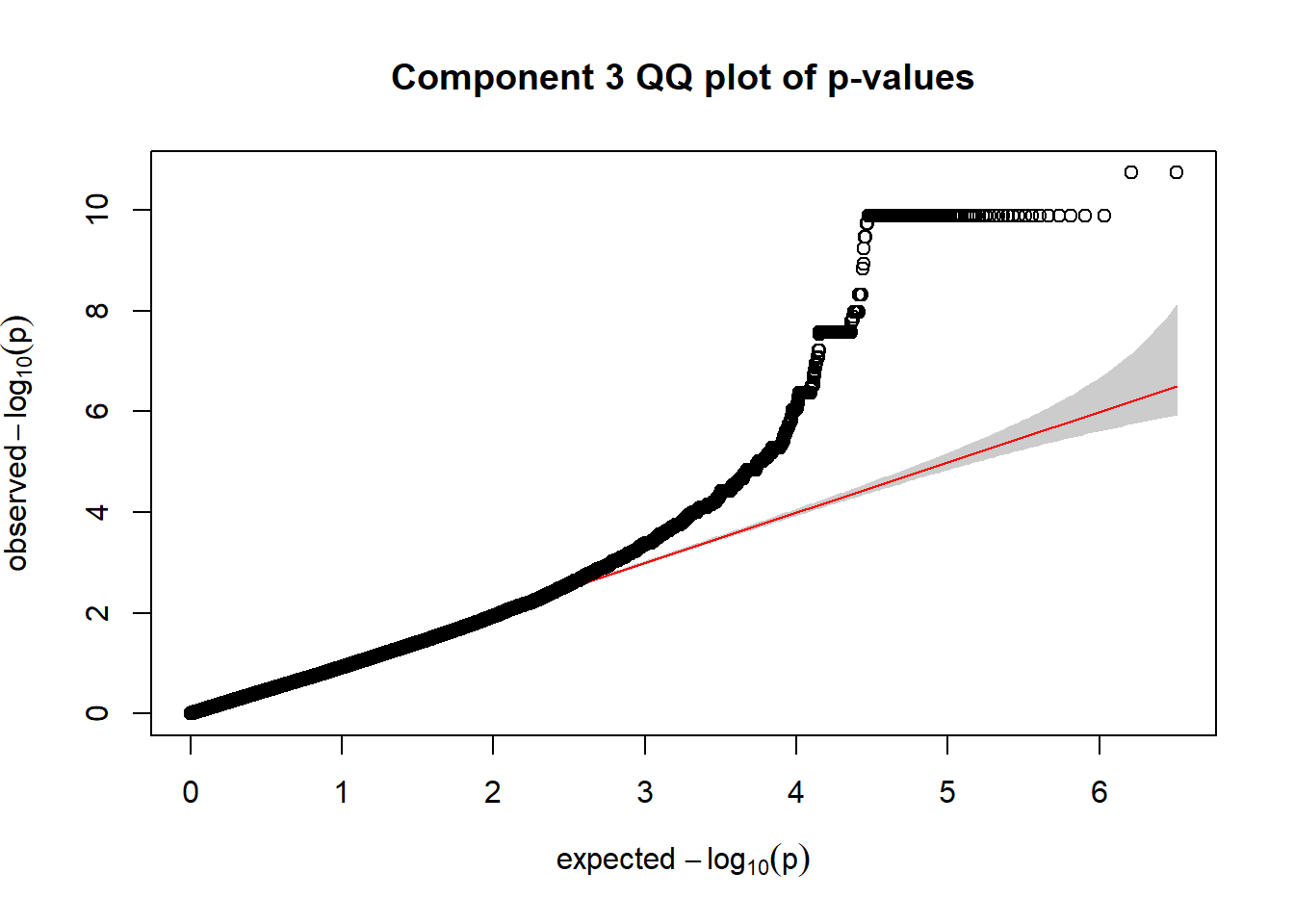
For component 3 (Wald)
qqplot.pvalues(pc3_wald$p)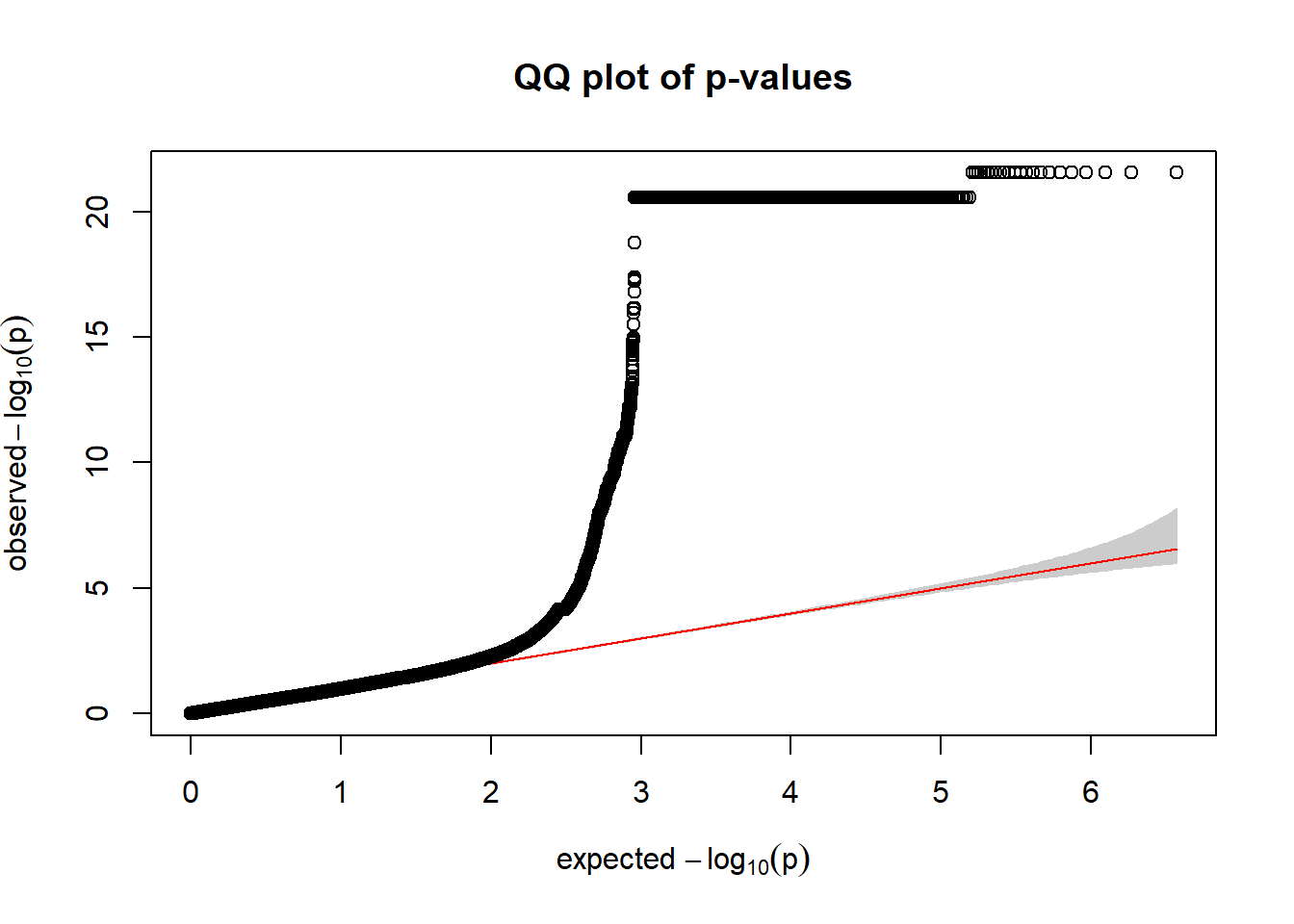
For R K-value V
qqplot.pvalues(r_kvalV_gwas$p)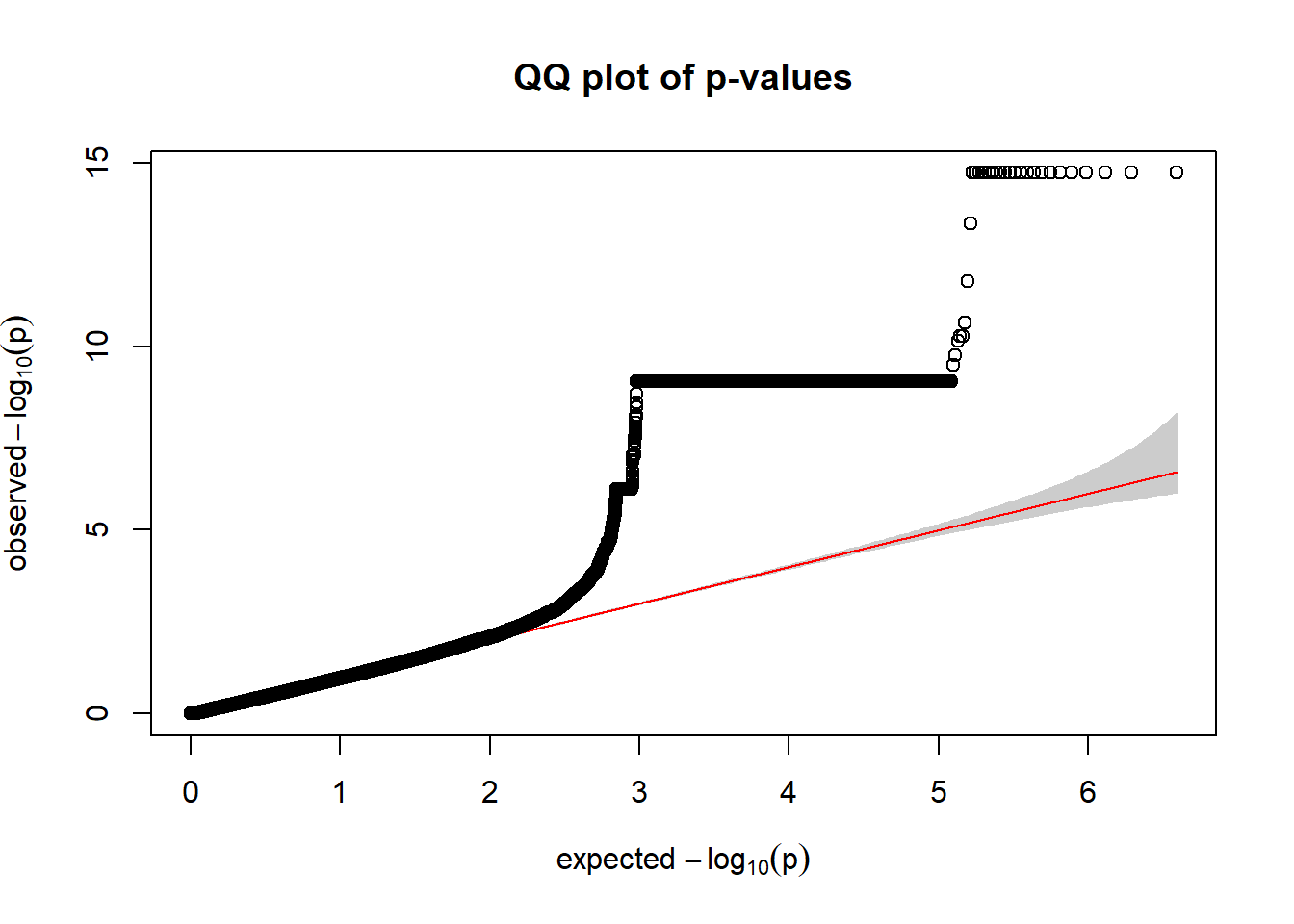
For R Kvalue V (wald)
qqplot.pvalues(r_kvalV_wald$p)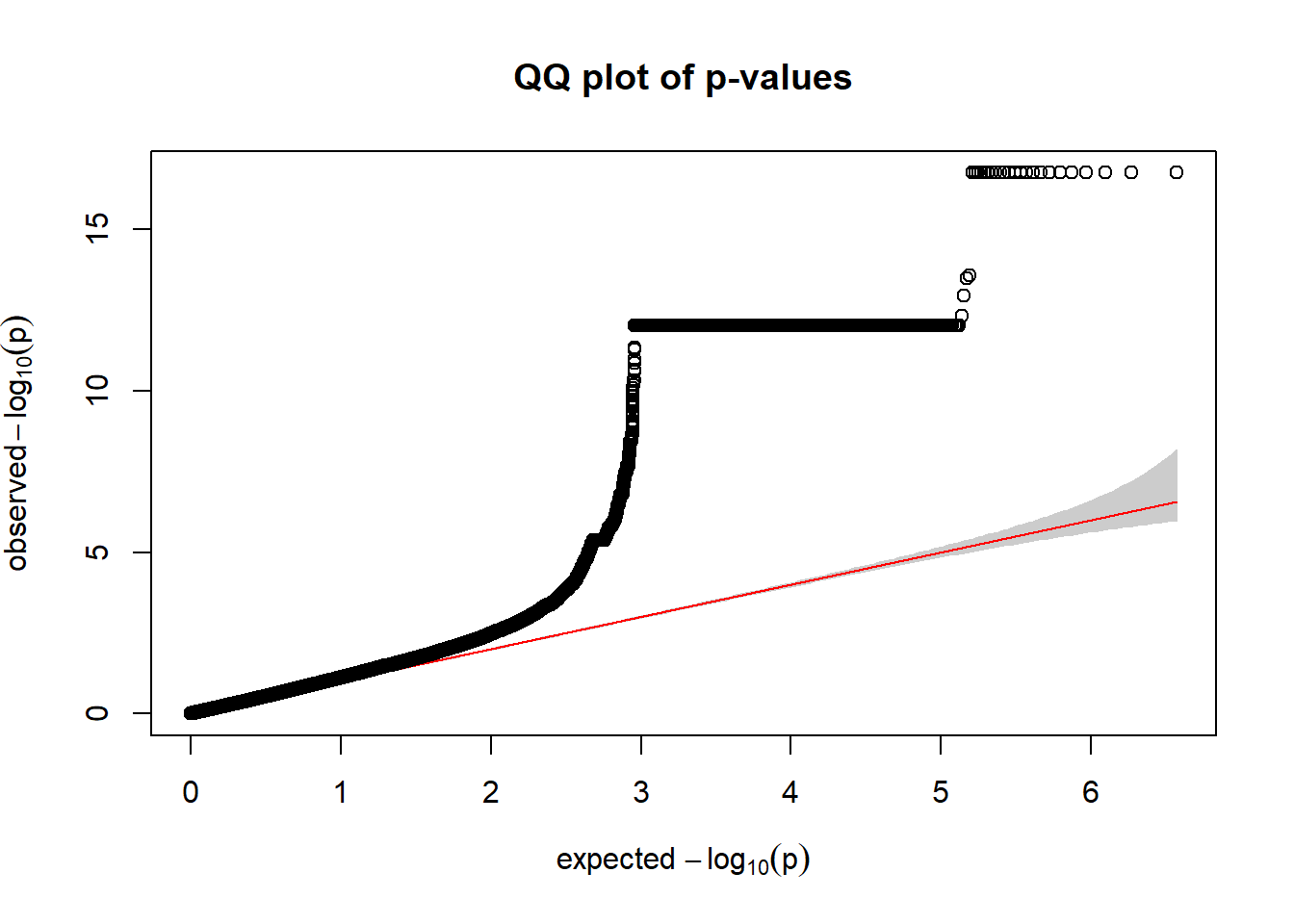
l_kvalV_gwas <- association.test(merged_nies_210818, nies_heritable_pheno$L.K.value.V, method="lmm", test = "lrt", K = merged_nies_GRM, eigenK = merged_nies_eiK, p = 2)
l_kvalV_gwas <- na.omit(l_kvalV_gwas)
l_kvalV_filtered <- l_kvalV_gwas %>% filter(freqA2 < 0.99)
qqplot.pvalues(l_kvalV_filtered)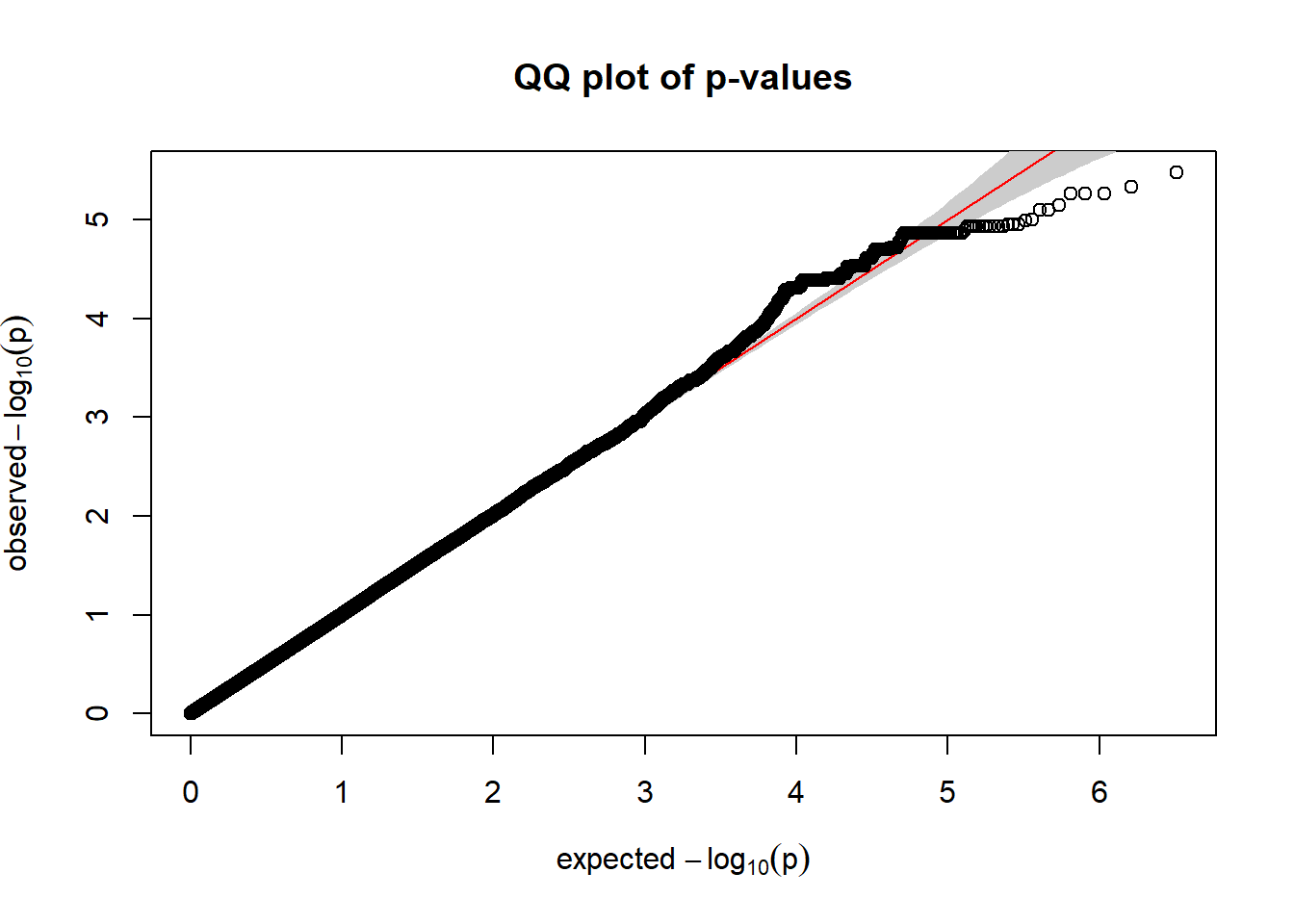
6. Generate QQ plots for all manhattan plots regardless of presence of significant loci
pheno_qqplot <- NULL
for (i in c(2:ncol(nies_heritable_pheno))){
pheno_colnames <- colnames(nies_heritable_pheno[i])
her_pheno_gwas <- association.test(merged_nies_210818, nies_heritable_pheno[,i], method = "lmm",
test = "lrt", K = merged_nies_GRM, eigenK = merged_nies_eiK, p =2)
her_pheno_gwas <- na.omit(her_pheno_gwas)
pheno_gwas_filtered <- her_pheno_gwas %>% filter(freqA2 < 0.99)
tiff(paste('C:/Users/Martha/Documents/Honours/Project/honours.project/Data/gwas_plots/', pheno_colnames, '_qqplot.png'), width = 800, height = 500, res = 150, compression = 'lzw')
gaston:: qqplot.pvalues(pheno_gwas_filtered$p, main = paste(pheno_colnames))
dev.off()
}